Dry Lab Modeling
Drylab, one of the three core teams of Westview iGEM, is responsible for the computational aspect of the research project. This year, our team simulated and modeled the behavior of PHB depolymerase to determine its structural stability and proclivity toward mutation. We examined the Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF) of a PHB depolymerase wild type and a mutant that was subject to alanine scanning. We also conducted a docking study of both. This analysis revealed their probable binding sites, showed the protein’s flexibility, and enabled us to find each protein’s stability. Drylab members also learned the basics of CSS, HTML, and Java to contribute to the team website.
What is Poly(3-Hydroxybutyrate) (PHB) plastic?
Poly(3-Hydroxybutyrate) (PHB), also known as P3HB, is a naturally produced biopolymer made by bacteria or organisms as a way to store energy inside their cells. PHB belongs to polyhydroxyalkanoates (PHAs), a sustainable and biodegradable alternative to conventional plastics.
Chemically, PHB is a polyester of 3-hydroxybutyric acid units. It’s fully biodegradable and biocompatible—meaning it can break down safely in the environment or even interact with living tissue. Microorganisms degrade PHB into water, carbon dioxide, and biomass, leaving no microplastic waste.
PHB has mechanical and thermal properties similar to polypropylene (PP), allowing molding, extrusion, or fiber formation. It can be brittle, so research aims to improve it through blending or copolymerization.
PHB is produced by microbes under nutrient-limited conditions, such as low nitrogen but high carbon. Modern research explores engineered bacteria and photosynthetic production from CO₂.
Overall, PHB is a promising step toward sustainable plastics—renewable, biodegradable, and versatile.
What is PHB depolymerase?
PHB depolymerase is an enzyme found in bacteria that degrades polyhydroxyalkanoates (PHAs), enabling microbes to use PHB as a carbon source when nutrients are scarce (Jendrossek & Handrick, 2002).
It hydrolyzes PHB into D-β-hydroxybutyric acid (3-hydroxybutyrate), which is further converted to acetoacetate by 3-hydroxybutyrate dehydrogenase.
Current research focuses on improving its activity and stability for efficient PHB degradation and waste management applications.
Alphafold Generated Model of PHB Depolymerase
(Jumper et al., 2021)
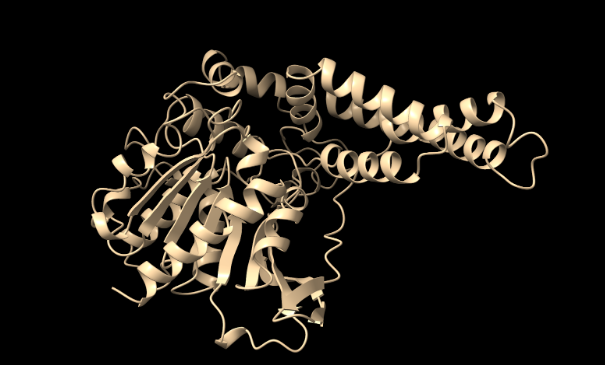
Figure 1. Predicted 3D structure of PHB depolymerase generated using AlphaFold (best of 5 models, 6 recycles, with relaxation).
FASTA Sequence:
MLYSLVEMNRAAMAPLRLTAKAASTFWGSPANPASSSQFARSITAGAKVFERATRYYGKPDWQIETTEIDGLPHAVTPNTVWGSPWCQLVRFDIEGVDTESRPDLIIVAPLSGHYATLLRGTVEAFLPTHNVHITDWTDARMAPVWLGRFDLDDYIDHIRDMLGFLGAGAHVLAVCQPGPPVLAAISMMAEDGSASLPATMTFMGSPIDARRSPTVPNKLAEERSLDWFRSKMIHSVPLPWPGAGRSVYPGFIQLSSFMHMNWDRHVDAQARFFDHLVEGDEDGADKHKTFYDEYLSVLDLTEEFYIQTIERVFQKALLARGEYRYRDERLVKPQAIKTVGLMTVEGELDDISGIGQTQAAHDLCTGIPGKFKEDYVQPGVGHYGVFNGSKFRQQIVPRVIAFQNHVTKG
What is 3-HB dehydrogenase?
3-Hydroxybutyrate dehydrogenase (3-HBDH) catalyzes the reversible reaction:
Acetoacetate + NADH + H⁺ ⇌ 3-Hydroxybutyrate + NAD⁺
This enzyme plays a role in both microbial and human energy metabolism. In bacteria, it helps convert acetoacetate into 3-HB, feeding into PHB biosynthesis.
Alphafold Generated Model of 3HV*-Dehydrogenase
(Jumper et al., 2021)
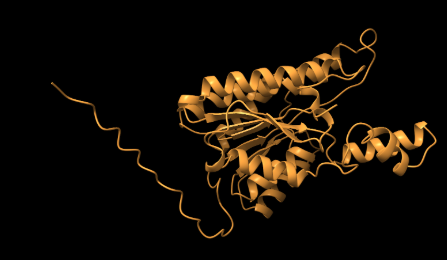
Figure 2. Predicted 3D structure of 3HV*-dehydrogenase generated using AlphaFold (best of 5 models, 6 recycles, with relaxation).
*3HV-dehydrogenase is a specialized version of 3-HB dehydrogenase.
FASTA Sequence:
MLFARLVLNKASLPLKNYGPSMLHRGKTALVTGSTSGIGKAIAEALAAEGANIVLNGLTKPGEDEALVADFEKRFDVKVGFSGANLTDPEAIEGLMGYAARDFGGVDILVNNAGVQHVSPIEDFPVAKWDLIIALNLSAAFHTTRMAMAGMKEKGWGRIINTASAHALVASPYKAAYVAAKHGIAGLTKTVALEGAQHGVRCNAICPGYVHTPLVDAQIADTAKARGISEEDVVNDVILAAQPTKEFVTVEQIGAMAVFLTSPAANQVNGALLQMDGGWVAQ
Usefulness of Mutations
Alanine scanning and other mutations help identify residues essential to enzyme function. Comparing mutant and wild-type proteins reveals which residues contribute to stability, catalysis, and substrate binding.
Mutations can improve enzyme performance under industrial conditions, such as heat tolerance and catalytic efficiency.
References
- Jendrossek, D., & Handrick, R. (2002). Microbial degradation of polyhydroxyalkanoates. Applied Microbiology and Biotechnology, 61(1), 94–108. https://pmc.ncbi.nlm.nih.gov/articles/PMC4664802/
- Jumper, J., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873), 583–589. https://doi.org/10.1038/s41586-021-03819-2
- ScienceDirect. (n.d.). Poly(3-hydroxybutyrate) (PHB). Retrieved October 7, 2025, from https://www.sciencedirect.com/topics/chemistry/poly-3-hydroxybutyrate
- Zhang, J., Cai, H., Song, J., & Chen, G. (2022). Structural and functional characterization of 3-hydroxybutyrate dehydrogenase involved in microbial metabolism. Frontiers in Microbiology, 13, Article 948369. https://www.frontiersin.org/articles/10.3389/fmicb.2022.948369/full
Molecular Dynamics Introduction
Our project used molecular dynamics (MD) simulations to investigate PHB depolymerase behavior. Below are the methods, computational setup, and results summary.
Methods
Simulations were run in OpenMM with GPU acceleration. Systems used the AMBER14 force field, TIP3P water, and physiological salt (0.15 M NaCl).
Energy minimization, heating, NPT equilibration, and production were performed sequentially. Each step ensured stability and reproducibility through checkpoints and custom NaN checks.
Summary
OpenMM applied Newton’s laws to each atom (Fi = -∇iU), updating every 2 fs to capture fine motions. The trajectories provided RMSD and RMSF data for enzyme flexibility and binding analysis.
Simulation Setup
- PDB structure files for the enzyme
- Python scripts with parameters
- AMBER14/TIP3P force field files
- Checkpoint files for restarts
This setup yielded trajectories describing PHB depolymerase’s structural dynamics at atomic detail.