Cycle 1: Investigate the main hazards of microplastics to the human
body
(1) Research
Initially, we assumed that microplastics mainly caused physical damage. However, through
research and preliminary experiments, it was discovered that oxidative stress and
inflammatory responses were the key pathogenic mechanisms. This refuted our initial
assumption of only considering physical damage. Therefore, we turned to intervention at
the molecular mechanism level. We investigated the most common harmful diseases caused
by microplastics in the human intestinal tract. Currently, PET has not found any
diseases that are significantly related to specific diseases. Among them, inflammatory
bowel disease (IBD) has the most active research on the association with microplastic
exposure and has been proven to have a certain correlation. Additionally, microplastics
can also cause dysregulation of the intestinal microbiota and damage to the intestinal
barrier function[1-3].
(2) Imagine
In addition to literature review, we aim to identify the targets of microplastics
causing intestinal diseases through bioinformatics methods for the purpose of designing
treatments. We plan to start with IBD, a common disease.
(3) Design and Build
The students in the modeling group came up with specific plans for target
identification:
a. Data collection and intersection with targets.
b. Construction of protein networks and screening of core nodes.
c. Multi-dimensional functional enrichment and verification.
d. Final determination through machine learning algorithms.
The specific methods are presented in the Test section.
(5) Data collection and intersection with the target
Firstly, the researchers established two independent gene databases. One was for PET
microplastics, where potential targets that might interact with it were comprehensively
collected from multiple databases such as PubChem, CHEMBL, and STITCH based on the
SMILES chemical encoding of its main degradation products. The other was for
inflammatory bowel disease (IBD), where a group of genes with the highest association
with IBD were selected from disease databases such as GeneCards and OMIM. By taking the
intersection of these two large databases, the "common targets" that were related to
both PET microplastics and IBD were finally identified. This step ensured that the
targets for the subsequent analysis had dual correlations.

Fig 1. The search for cross-targets between PET and IBD
(6) Protein network construction and core node selection
Just having a list of genes is not enough, because genes usually work together through
complex networks. Researchers used the STRING database to construct a protein-protein
interaction (PPI) network based on the common targets from the previous step.
Subsequently, this network was imported into the Cytoscape software for visual analysis.
The researchers employed two complementary algorithms to identify the key genes in the
network:
a. CytoHubba_MCC algorithm: This is used to find the nodes with the most connections
and located in the "central hub" position. This method can directly identify the
proteins that play a key role in information transmission.
b. MCODE plugin: This is used to discover the most densely connected "functional
modules" or protein clusters in the network. This helps to identify the protein groups
that collaborate to perform specific functions in the cell.

Fig 2. Protein interaction network results
(7) Multidimensional functional enrichment and validation
To understand the specific biological functions of these core genes, the researchers
conducted Gene Ontology (GO) and KEGG pathway enrichment analyses. The results showed
that these genes were highly enriched in biological processes such as "inflammatory
response" and "immune response", which are highly relevant to the pathology of IBD, and
were closely related to important signaling pathways such as PI3K-Akt and Jak-STAT.

Fig 3. Metabolic pathway analysis
To verify whether PET can directly act on these targets at the physical level, we
conducted molecular docking and molecular dynamics simulation tests. The results showed
that the binding energies of PET to proteins such as JAK2, IL2, and TGFBR2 were
negative, which proved in physics that there is a stable and spontaneous binding between
them.

Fig 4. PET and three target molecules docking
(8) The machine learning algorithm is finally locked
After going through the above multiple rounds of screening and verification, a series of
machine learning algorithms (including LASSO regression, SVM-RFE, boruta and XGBoost)
were finally employed for the final and most rigorous screening. These algorithms
further evaluated the importance of each gene from a statistical perspective, and
ultimately precisely identified the three core targets: JAK2, IL2 and TGFBR2.

Fig 5. Machine learning screening
(9) Localization of pathogenic function of the target
JAK2: It belongs to non-receptor tyrosine kinases. The accumulation of ROS can directly
activate JAK2, making it an important upstream trigger point in the inflammatory
cascade.
IL2: T-cell growth factor, over-activation is associated with immune dysregulation and
abnormal T-cell proliferation in IBD.
TGFBR2: A key receptor of the TGF-β signaling pathway, regulating cell proliferation,
differentiation, and tissue repair. Its abnormality may lead to failure of intestinal
epithelial repair or a tendency towards fibrosis.
(1) Learn
We have identified three therapeutic targets for the harm caused by microplastics to the
human body. In the future, when we are searching for therapeutic components, we will pay
particular attention to their relationship with the target sites.
Cycle 2: Search for therapeutic elements - Antioxidants
(2) Research
Firstly, we address the issue from the first aspect of microplastics causing disease -
oxidative stress. Research indicates that reactive oxygen species (ROS) play a
significant role in the pathogenesis of inflammatory bowel disease. Superoxide anion
(O₂⁻) and hydrogen peroxide (H₂O₂) are two core types of ROS. The accumulation of ROS
can directly activate the JAK2 pathway and trigger an inflammatory cascade reaction.
Therefore, eliminating ROS is a key strategy for inhibiting inflammation induced by
microplastics.
(3) Imagine
Can we establish a "cohesive" antioxidant regulatory system to rapidly eliminate ROS at
the source and achieve antioxidant stress response? Does the molecular mechanism layer
have any effect on the targets we are exploring?
(4) Design and Build
There are two key enzymes that play a dominant role in the removal of ROS:
a. SOD1: Converts O₂⁻ into H₂O₂;
b. CTT1: Further decomposes H₂O₂ into water and oxygen.
Theoretically, the cascade reaction formed by these two enzymes can efficiently remove
ROS.

Fig 6. The function of the antioxidant module
(5) Test
After reviewing the literature, SOD1 and CTT1 were found to be capable of performing
their intended functions in our chassis - Saccharomyces cerevisiae. Therefore, we
selected these two genes as the effect modules and designed a dual-enzyme expression
pathway to enable the engineered bacteria to possess systematic antioxidant
capabilities.
In terms of the molecular mechanism, this antioxidant component directly eliminates
reactive oxygen species (ROS) to block the activation of JAK2 at the source, achieving
direct intervention in the downstream inflammation[22][23].
(6) Learn
The team realized that the antioxidant module can precisely address the "downstream
trigger point" of ROS → JAK2, but the inflammatory network is complex, and relying on a
single module is not sufficient to completely inhibit the pathological process.
Oxidative stress is merely a "trigger", not the sole effector. Once the inflammatory
pathway is activated, merely removing the trigger factor is not enough to stop the
pathological progression. This discovery prompted the project to shift to a more
comprehensive treatment approach - in addition to the antioxidant module, functional
modules such as immune regulation and tissue repair need to be added.
Furthermore, model analysis and literature comparison indicate that this system can
indeed significantly clear ROS and directly block the activation of JAK2 in theory.
However, there is a fundamental defect: it can only eliminate the initial oxidative
signal, but cannot inhibit the downstream inflammatory cascade reaction triggered by
ROS. This deserves our team to continue to think about optimization strategies.
Cycle 3: Search for therapeutic elements - anti-inflammatory
(1) Research
The team first realized that direct intervention in the inflammation itself was
necessary to solve the problem. Thus, they embarked on in-depth research on the core
signaling pathways of the host immune system (such as
JAK2, IL-2, and TGFBR2). By analyzing the mechanisms of these
pathways, the team realized that they were key nodes regulating the immune system, and
theoretically, the inflammation could be controlled through external factors.
(2) Imagine
Based on this understanding, the team proposed an ambitious idea:
Utilizing the synthetic biology pathways of engineered bacteria, to secrete inhibitors
or agonists, to "remote control" the functions of the host immune cells from the
outside, directly acting on the JAK2, IL-2 and TGFBR2 pathways, in order to achieve
precise control of inflammation.
(3) Design and Build
During the specific design and analysis phase, the team decisively rejected this
approach. The reason was as follows: These host immune pathways are extremely
complex and mainly function within the host immune cells; engineered
microorganisms (such as yeast) are difficult to stably, safely and precisely directly
regulate them; directly interfering with key kinases such as JAK2 might cause
unforeseeable off-target effects on systemic functions such as hematopoiesis and
immune surveillance; and there are significant risks in terms of biological
safety.
Therefore, the team shifted its focus from "Is it technically possible?" to "Is it
functionally safe and controllable?". This strategic retreat avoided the high-risk dead
end. Subsequently, the team shifted to designing new solutions, seeking molecules that
could the organization's repair and stabilization recovery. Eventually,
Trefoil Factor 3 (TFF3) was identified as the ideal effector factor.
(4) Test
The team further analyzed the mechanism of action of TFF3:
Promoting epithelial repair: Activating the JAK2/STAT3 pathway in intestinal epithelial
cells in a local manner, promoting mucosal healing and recovery of barrier function.
Actively inhibiting inflammation: Reducing key pro-inflammatory cytokines (such as
TNF-α, IL-1β) in the inflammatory microenvironment, and weakening the excessive
activation of IL-2 and TGFBR2-related signals.
This dual function ensures both local and controllable efficacy, meeting safety
requirements[6].
(5) Learn
Through this cycle, the team learned that:
Direct intervention in the core host immune pathways is an unbridgeable red line because
it is unpredictable and high-risk;
Strategically giving up high-risk solutions and instead adopting more indirect and safer
mechanisms reflects engineering wisdom and a high sense of responsibility for biological
safety;
Ultimately, the team established the "antioxidation + mucosal repair" strategy to
replace the direct blocking of inflammation, thereby achieving the anti-inflammatory
goal.
Cycle 4: ROS-Responsive Promoter Design
(1) Research
In the inflammatory environment of the human intestinal tract, a large amount of
reactive oxygen species (ROS) accumulate, driving epithelial damage and inflammatory
cascade reactions. We need a regulatory element that can precisely sense ROS signals as
a molecular switch for the therapeutic pathway, so as to achieve the expression of
conditionally induced anti-inflammatory factors.
(2) Imagine
The literature indicates that the transcription factors Yap1 and Skn7 in Saccharomyces
cerevisiae are key regulatory elements for sensing ROS:
Yap1: Highly sensitive to ROS such as hydrogen peroxide, and can serve as the main
sensing module;
Skn7: Provides an auxiliary function, and can enhance the stability and intensity of the
response;Synergistic effect: When Yap1 and Skn7 synergistically bind to the promoter,
they can significantly increase the gene expression level and accelerate the response
speed.
Therefore, we hypothesize that the natural ROS-responsive promoter regulated by the
synergistic control of Yap1 and Skn7 can be used as a candidate regulatory tool.
(3) Design and Build
Based on the regulatory mechanism of Yap1/Skn7, we have identified three types of
naturally existing promoters in yeast:
pGSH1 (glutathione synthetase promoter): contains multiple Yap1 binding sites;
pTRX2 (thioredoxin reductase promoter): contains Yap1 sites and Skn7 elements;
pGPX2 (glutathione peroxidase promoter): contains Yap1 sites and Skn7 elements, and can
simultaneously sense ROS and inflammation-related oxidative signals.
These candidate promoters can all respond to oxidative stress signals, but there are
differences in their activation intensity, response speed, and background leakage.
We conducted a literature review and comparative analysis of the three types of
promoters[25]:

Table 1. Oxidative stress promoter selection
After comprehensive analysis, pGPX2 stood out:
Low background expression → Avoid energy waste and side effects during non-inflammatory
states;
High induction intensity → Capable of rapidly producing sufficient therapeutic proteins
during inflammatory outbreaks;
Fast response → Exhibits a faster response speed to ROS and inflammatory signals
compared to other promoters;
Broad response spectrum → Covers ROS and a wider range of inflammatory-related oxidative
signals.
Therefore, the pGPX2 promoter has been confirmed as the optimal ROS-responsive
regulatory tool, constituting the core component of our treatment line design.
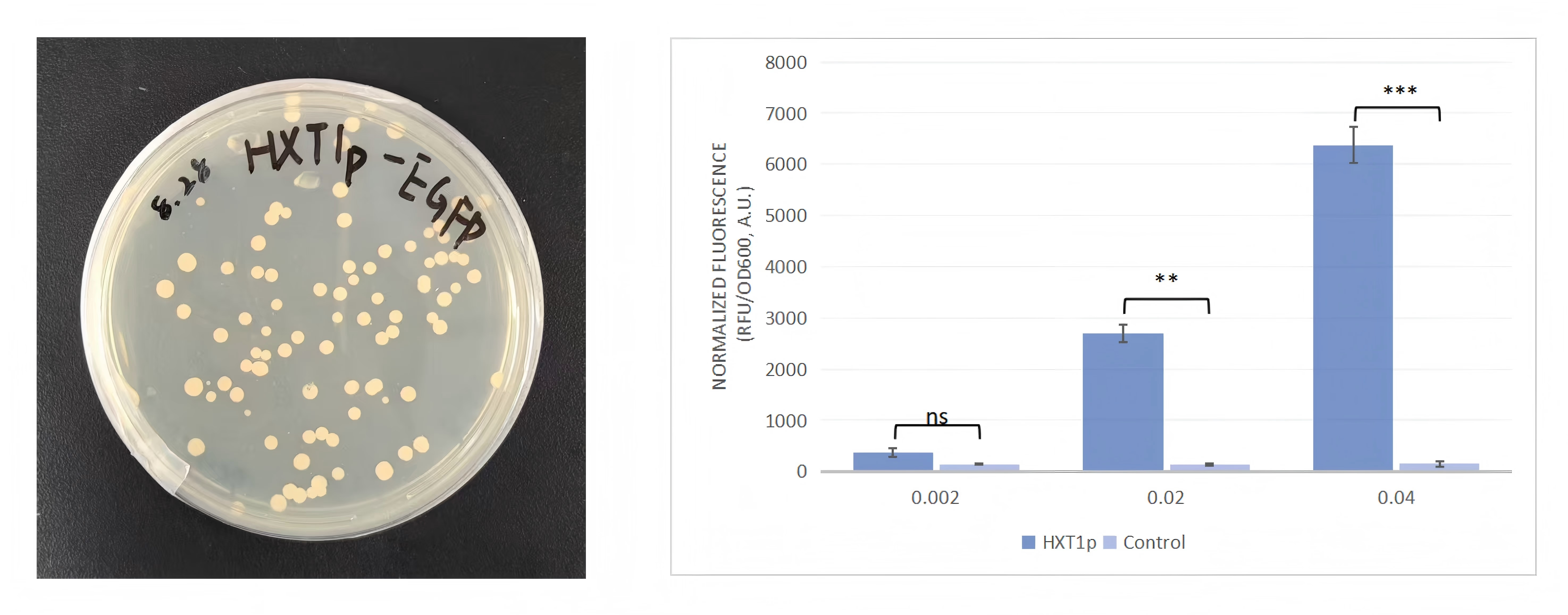
Based on the literature, we found the pGPX2 promoter from the yeast genome, located
upstream of the GPX2 CDS fragment (-709/+7).

Fig 7. GPX2 promoter gene map
(4) Learn
We confirmed that pGPX2 is the optimal ROS-responsive promoter, capable of achieving low
background, high intensity, and rapid response characteristics in an inflammatory
environment, providing a reliable "molecular switch" for the therapeutic pathway.
However, in the subsequent design process, when we attempted to place SOD1, CTT1, and
TFF3 under three separate pGPX2 promoters respectively, new challenges emerged:
Metabolic burden: Multiple parallel pathways would significantly increase energy
consumption;
Expression incoordination: Even relying on the same promoters, the dynamics of the three
pathways are difficult to be completely consistent;
Uncontrollable dose ratio: The production of the three proteins is difficult to maintain
a stable stoichiometric ratio, thereby weakening the synergistic effect.
Therefore, the team realized that the selection of a single high-quality promoter is
only the first step. The more crucial issue lies in how to achieve safe, efficient, and
coordinated expression of multiple proteins within the same pathway. This thinking
directly led to the next round of iteration (Cycle 5): how to achieve the joint control
of the anti-inflammatory and antioxidant modules.
Cycle 5: How can the joint control of anti-inflammation and
anti-oxidation be achieved
(1) Research
After identifying the final three effect proteins, SOD1, CTT1 and TFF3, the team needed
to address a key issue:
How to rationally express these proteins in the engineered bacteria to maximize their
synergistic effect?
The research found that if each protein was driven by an independent expression pathway,
it would cause excessive metabolic burden; if the expression was not synchronized, it
would be difficult to ensure the precise stoichiometric ratio; and this would further
weaken the synergistic therapeutic effect of the three proteins.
(2) Imagine
Based on this, the team envisioned giving up the "parallel scheme" of multiple
independent lines and instead attempting a more efficient and integrated approach:
Integrate multiple effector proteins into a single line to ensure their synchronous
expression, thereby enhancing the robustness and safety of the system.
(3) Design and Build
The team finally constructed a single-line multi-module design: GPX2 → NCW2-SOD1 → P2A →
NCW2-TFF3 → T2A → NCW2-CTT1.
The key design points include:
Promoter (GPX2): Only drives expression when inflammatory signals are present, ensuring
the synchrony and spatiotemporal consistency of the three proteins.
2A self-cleavage peptide (P2A, T2A): Initially, an attempt was made to fuse the three
proteins through a flexible linker, but analysis indicated that it would disrupt their
three-dimensional structure and lead to functional loss. The team conducted literature
research and adopted 2A self-cleavage peptides, enabling a single mRNA to generate three
functionally independent proteins in a 1:1:1 ratio during translation, maintaining
structural integrity while reducing metabolic burden.

Fig 8. The mechanism of action of 2A peptide[8]
Secretion signal peptide (NCW2): Add the NCW2 sequence before each protein, directing
the product to be secreted outside the cell and ensuring its release as needed in the
intestinal lumen[9].
By combining various elements, we have developed our treatment plan.

Fig 9. Treatment Module Diagram
(4) Test
In the scheme simulation and literature comparison, the team verified that the single
promoter + 2A peptide strategy is feasible and has been widely applied in multi-protein
systems; the 2A peptide can ensure a stable stoichiometric ratio and avoid functional
loss caused by the fusion protein; the application of the secretion signal peptide NCW2
has been well reported in eukaryotic expression systems, and can reliably guide protein
secretion.

Fig 10. Comparison of signal peptide efficiency[9]
Influences on heterologous protein secretion by accessory proteins linked to SPs with
different properties. a) α-amylase secretion titer in the ref, △SPC1 and ΔSEC72 strains
when using different SPs. The α-factor SP was used as a reference to compared with other
11 super-secreted SPs. b) Hydrophobicity of SPs determined α-amylase secretion change
tendency (increase or decrease) in the ΔSEC72 strain. The secretion change was
calculated as follow: (amylase secretion by the ΔSEC72 strain – amylase secretion by the
ref strain)/(amylase secretion by the ref strain) × 100%. was used as the control
strain. Data shown are mean values ±SDs of duplicates[27-28].
After the information was confirmed, we began the wet experiment work, which was divided
into two parts: the construction of ΔSEC72 Saccharomyces cerevisiae and the construction
and introduction of therapeutic genes. Preliminary laboratory results have been obtained
at present.

Fig 11. Treatment module-related gene map
(5) Learn
Through this round of cycle, the team realized that a single-line multi-module design
can significantly reduce genetic complexity and metabolic burden; using 2A self-cleaving
peptides is the most elegant way to solve the problem of multi-protein co-expression,
which is both efficient and reliable; in the iGEM project, engineering wisdom lies not
only in "getting the proteins expressed", but also in how to express them safely,
efficiently and coordinately to maximize the therapeutic synergy.
Cycle 6: The auxiliary role of symbiotic systems in treatment
(1) Research
After modeling and testing the engineered yeast with the integrated SOD1–TFF3–CTT1 gene
circuit, we found that the performance of the single yeast treatment module still had
limitations: although it had significant effects on ROS clearance (targeting the
upstream signal of JAK2) and intestinal barrier repair (affecting the sustained
activation of IL2), it was still insufficient in terms of the breadth of immune
regulation and long-term stability maintenance. Moreover, from the literature
comparison, it was also clear that there was a knowledge gap: this module had no known
direct connection with the TGFBR2 signaling pathway.
(2) Imagine
In response to this performance gap, the team did not choose to continue stacking more
complex circuits in yeast. Instead, they proposed a new idea: introducing a naturally
immune-regulating partner that can complement the yeast; expanding the engineering goal
from a single cell to a multi-microbial system, achieving a transition from molecular
engineering to micro-ecosystem engineering.
(3) Design and Build
The design concept is to combine the precise elimination ability of engineered yeast
with the broad-spectrum regulatory ability of probiotics to achieve coordinated
intervention at both the upstream and downstream of the inflammatory signals.
a. The molecular circuitry of the yeast treatment module (precise elimination and
repair)
The core of the engineered yeast is the condition-inducible therapeutic pathway, as
described above: pGPX2 → NCW2-SOD1 → P2A → NCW2-TFF3 → T2A → NCW2-CTT1.
b. The natural immune regulation advantages of DT88 (broad-spectrum regulation and
signal weakening)
The Lactobacillus plantarum DT88, as an auxiliary strain, compensates for the
deficiencies of the yeast module at the ecological and signaling levels through its
natural metabolic and cytokine regulatory capabilities:
Inhibiting pro-inflammatory factors:DT88 can significantly reduce the elevated
levels of pro-inflammatory cytokines TNF-α, IL-6, and IL-1β in the microplastic-exposed
mouse model, while simultaneously synergistically enhancing the production of
anti-inflammatory factor IL-10.
Inhibiting co-stimulatory molecules: The symbiotic system has been proven to be
able to reduce the expression of activation markers CD86 on the surface of monocytes.
Metabolic products anti-inflammatory: The short-chain fatty acids (SCFAs)
produced by DT88, especially butyric acid, can inhibit the NF-κB signaling pathway in
immune cells and epithelial cells, reducing the transcription of inflammatory factors at
the nuclear level.
(4) Test
The objective of this section is to combine the evidence from the literature and the
modeling results to verify whether the dual intervention of the symbiotic system can
precisely act on the core target, and to focus on evaluating the micro-regulatory effect
of DT88.
a. Immunomodulatory effects:
The co-culture system of Saccharomyces cerevisiae and Lactobacillus exhibits unique
immunomodulatory properties, and its anti-inflammatory effect is significantly superior
to that of a single strain[10]. This synergy is not simply additive; rather,
it
stems
from the host immune system's specific recognition and response to the combined patterns
of microbial molecules (MAMPs). In mice exposed to microplastics, DT88 can restore
elevated levels of TNF-α, IL-6, and IL-1β to normal, and promote an increase in IL-10,
thereby guiding the immune response towards a tolerative state[11].
b. Inhibition of immune cell activation:
This system can significantly reduce the expression of activation markers CD14 and CD86
on the surface of monocytes, demonstrating its stable effect in inhibiting excessive
inflammatory signals[10].
c. Contribution to metabolic homeostasis:
Microbial metabolites, short-chain fatty acids (SCFAs), are the main energy source for
colonic epithelial cells, promoting their proliferation and repair. At the same time,
SCFAs can inhibit the NF-κB pathway, reduce the release of inflammatory factors, and
stimulate goblet cells to secrete mucin to strengthen the intestinal mucus
barrier[12].
d. Molecular targeting mechanism:
JAK2 pathway (upstream weakening): DT88 reduces the levels of TNF-α and IL-6, thereby
indirectly inhibiting the activation of the JAK/STAT pathway and synergistically
promoting the production of IL-10.
IL-2 pathway (co-stimulation inhibition): This system can down-regulate the expression
of CD86 on monocytes, which is equivalent to adding a "molecular brake" in the T cell
activation circuit, alleviating the excessive immune activation induced by
microplastics.
TGFBR2 pathway: Current research shows that there is no known direct molecular
association between this system and TGFBR2.

Table 2. The effect of the symbiotic system on the target
(5) Learn
In conclusion, the therapeutic effect of this symbiotic system is multi-level and
three-dimensional. It has established a "deep defense" strategy. Additionally, we later
discovered that the symbiotic system has a reinforcing effect on the barrier structure:
The symbiotic system achieves structural reinforcement by regulating tight junction
proteins (TJPs). Exposure to microplastics can damage the integrity of the intestinal
barrier. Probiotics interact with epithelial cells and upregulate the expression of
"sealing-type" proteins such as ZO-1, Occludin, and Claudin-1, reducing the permeability
of the paracellular pathway and preventing harmful substances from leaking. Thus, this
module has initially solved the problem of the harm caused by microplastics to the human
body. In the next step, we need more experiments for verification[32].
In conclusion, the therapeutic effect of this symbiotic system is multi-level and
three-dimensional. It has established a "deep defense" strategy. Additionally, we later
discovered that the symbiotic system has a reinforcing effect on the barrier structure:
The symbiotic system achieves structural reinforcement by regulating tight junction
proteins (TJPs). Exposure to microplastics can damage the integrity of the intestinal
barrier. Probiotics interact with epithelial cells and upregulate the expression of
"sealing-type" proteins such as ZO-1, Occludin, and Claudin-1, reducing the permeability
of the paracellular pathway and preventing harmful substances from leaking. Thus, this
module has initially solved the problem of the harm caused by microplastics to the human
body. In the next step, we need more experiments for verification[13].