3.1 Experimental Images
3.1.1 Experimental Plating and Image Acquisition
The spread plate experiment takes E.coli
as the research object. The plating operation is completed on the aseptic
operating table in the laboratory. LB solid medium is used to support the
growth of bacterial colonies. After plating, the plates are inverted and placed
in a 37°C constant-temperature incubator for cultivation. During the
cultivation period, photos are taken at regular intervals to record the growth
of bacterial colonies, and the following precautions must be strictly followed
during shooting:
① The same camera shall be used for shooting and recording
throughout the process, and the shooting distance and storage format shall be
fixed to avoid the impact of differences in equipment parameters on subsequent
recognition.
② Shooting shall be conducted in an environment with uniform
light, without direct strong light. The lens shall be perpendicular to the
surface of the medium and fully cover the area of the Petri dish.
③ Place the Petri dish on a solid-color, pattern-free
background board to eliminate interferences such as background variegation and
shadows, ensuring that the outline of bacterial colonies is clearly
distinguishable.
④ Take the moment when the plate is placed into the
incubator as the "0h" starting point, take photos at irregular
intervals to record the growth of E.coli, and finally obtain 15-20
images covering the entire growth cycle.
⑤ Name the images according to the "cultivation
time" and record the shooting time to facilitate subsequent data
organization.
3.1.2 Image Processing
To eliminate differences in angle and field of view among
colony images taken at different times and ensure unified scales during
subsequent target detection, we perform standardization processing on the images.
Through methods such as rotation (adjusting the image angle to make the edge
direction of the medium consistent) and cropping (taking the edge of the medium
as a fixed control reference to crop out the excess background area), all
processed images have a unified scale and consistent visual starting point,
preventing shooting deviations from affecting the accuracy of colony feature
extraction. The processed images are shown in the figure below:

Figure 2: Processed Plate Images
The unit of the image names is "hours",
corresponding to the growth time of E.coli.
3.2 Intelligent Recognition
Intelligent recognition is the core link connecting
"standardized images" and "quantified colony features". Its
purpose is to extract key information that can be used for music translation,
such as the position, length, and width of colonies, from the processed E.coli
growth images. To avoid the efficiency bottlenecks and accuracy deviations
caused by manual operations, this module adopts the target detection technology
based on YOLOv8s to realize the automatic recognition of colony information,
providing stable and batch structured data support for subsequent data analysis
and music translation.
3.2.1 Data and Preprocessing
① Dataset
The dataset used for training in this module is derived from
the free part of the AGAR dataset [C1] and
the dataset provided in relevant literatures [C2]. A total of 140 images and corresponding data have
been sorted out, mainly involving five core colony categories: B.subtilis, C.albicans, E.coli,
P.aeruginosa, S.aureus, as well as a small number of mixed colonies.
We sincerely appreciate the efforts and contributions of relevant researchers
and staff.
② Format Conversion Processing
Subsequently, we need to perform format processing on the
obtained dataset. The process involves converting the original JSON annotations
into XML files in VOC format first, and then further converting them into txt
files dedicated to YOLO. While standardizing the bounding box coordinates into
normalized values required for model training, the automatic division of the
training set (80%) and validation set (20%) is completed, providing
standardized and directly readable input data for subsequent data processing
and efficient training of the YOLOv8s model.
③ Image Slicing and Algorithm Optimization Processing
We first conducted a training session using the existing
dataset, but we found that the training results had problems in recognizing
extremely small colonies in practical instances. Therefore, after converting
JSON to XML files, we implemented algorithm adaptability optimization through
image slicing processing. Large-sized or irregular images are converted into
sub-regions of fixed size, and label information is adjusted synchronously to
ensure that the colony features in the slices are complete and the label
coordinates are accurate.
We adopt the sliding window method for slicing: using a
preset fixed size as the window, calculating the sliding step according to the
set overlap rate, and successively intercepting sub-regions on the original
image; synchronously processing the VOC format labels, retaining only the
colony targets completely located within the window, converting their
coordinates into relative coordinates within the slices and generating new
labels; performing grayscale processing on some truncated colony regions to
weaken the features of incomplete targets and eliminate interferences; finally
outputting label files corresponding to the slices one-to-one to maintain the
consistency of the data structure, laying a foundation for the YOLOv8s model to
efficiently learn colony features.

Figure 3 : Effect Diagram of Slicing and Grayscale Masking
3.2.2 YOLOv8s Model
YOLOv8 is a major updated version based on YOLOv5, released
by Ultralytics in January 2023. On the basis of previous YOLO versions, it
introduces new functions to further improve the performance and flexibility of
the model. The following figure shows the overall framework of YOLOv8:

Figure 4: Model structure of YOLOv8 detection models from RangeKing@github[C3]
① Backbone
The Backbone of YOLOv8 is composed of three major modules
(Conv, C2f, and SPPF) combined in a logical manner. The Conv module is the
foundation, which realizes downsampling through the structure of "2D Convolution
+ BatchNorm2d + SiLU" to provide fixed scales for subsequent feature
extraction. C2f is a key improvement, which has fewer parameters and stronger
feature extraction capabilities than the C3 module of YOLOv5. Through the
process of "ConvModule Processing → Split into Two Paths → One Path
Directly Connected, the Other Path Passing Through DarknetBottleneck with
Residual → Concat Concatenation → ConvModule Output", it reduces the loss
of shallow features and captures fine-grained information. Consistent with
YOLOv5, SPPF fuses features through multi-scale pooling and retains global
semantics. Overall, it consists of 5 Conv modules, 4 C2f modules with different
parameters, and 1 SPPF module, forming a complete feature extraction chain from
underlying textures to high-level semantics.
② Neck
The Neck adopts a combined structure of "FPN +
PAN", whose core is to realize the bidirectional fusion of high-level and
low-level features. FPN (Feature Pyramid Network) works in a
"top-down" manner: after upsampling the high-level small-sized
feature maps of the Backbone, it sequentially concatenates them with the
middle-level and low-level feature maps, followed by C2f processing, to
transmit high-level semantic features and solve the problem of low-level
semantic ambiguity. PAN (Path Aggregation Network) operates in a
"bottom-up" way: after downsampling the low-level feature maps fused
by FPN, it sequentially concatenates them with the middle-level and high-level feature
maps, followed by C2f processing, to transmit low-level positioning features
and make up for the insufficient positioning of high-level features. Finally, 3
optimized feature maps corresponding to different scales are output.

Figure 6: Schematic Diagram of the Feature Pyramid Network Structure
③ Head
The Head adopts the innovative design of "Anchor-Free +
Decoupled-Head" to improve the flexibility and accuracy of prediction.
Anchor-Free does not require preset anchor boxes; it directly infers the
position and size of the target from the feature maps of the Neck, adapts to
changes in object size, and avoids missed detection and repeated detection
caused by unreasonable anchor boxes. The Decoupled-Head splits
"classification + regression" into independent branches, both of
which are processed through "4 3×3 convolutions + 2 1×1
convolutions". The classification branch uses BCE (Binary Cross-Entropy)
loss to optimize category judgment, and the regression branch uses CIOU
(Complete Intersection over Union), WIOU (Weighted Intersection over Union),
and DFL (Distribution Focal Loss) to optimize the bounding box coordinates.
Finally, the target box coordinates and category probabilities are output, and
the results are obtained through non-maximum suppression. Additionally, it
outputs 3 feature maps corresponding to the Neck, realizing accurate prediction
of targets of different sizes.

Figure 7: Schematic Diagram of the YOLOv8 Head Structure, from MMYOLO[C4]
④ Model Performance
The following figure shows the performance curves of
multiple YOLO series models:

Figure 8: YOLOv8 Performance Curve [C5]
Considering the size of our training set, we decided to
select YOLOv8s as our training model.
3.2.3 Training Results
Based on the preprocessed dataset and the YOLOv8s model, we conducted
300 rounds of training on 1421 images in the training set and 356 images in the
validation set. The results are as follows:

Figure 9: Statistical Distribution Visualization of the Colony Dataset
From the statistical distribution visualization of the dataset,
it can be seen that the sample sizes of the five core colony categories are
sufficient, providing rich feature learning materials for model training, which
is an important foundation for the model's excellent recognition performance on
the core categories. At the same time, the morphological characteristics of
colonies, such as random distribution in images and positive correlation
between width and height, also provide data-level support for model training.
These four figures are the F1-Confidence Curve,
Precision-Confidence Curve, Precision-Recall Curve, and Recall-Confidence Curve
respectively. Based on the comprehensive analysis of these four evaluation
curves, it can be seen that our trained model exhibits excellent colony
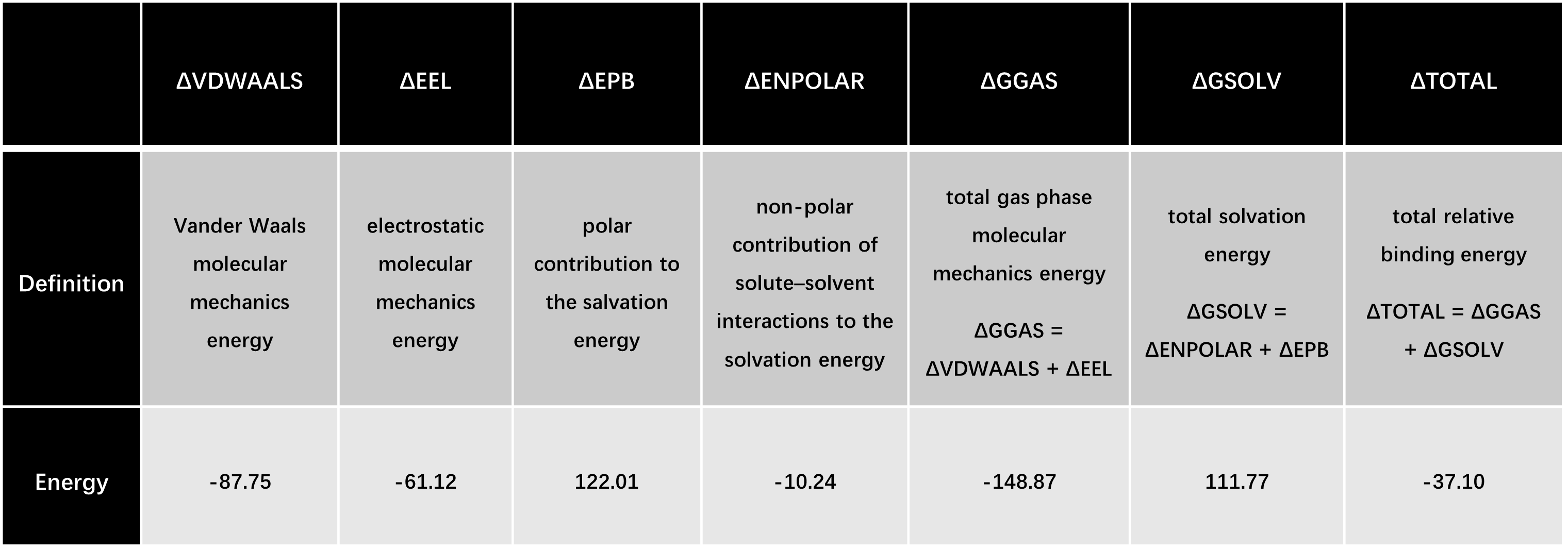
detection performance: in the Precision-Recall Curve, the overall mAP@0.5 (mean
Average Precision at IoU=0.5) is as high as 0.982, and the average precision of
core colonies such as B. subtilis, C. albicans, and S. aureus is close to 1.0,
indicating that the model has extremely high recognition accuracy for target
colonies. The Recall-Confidence and F1-Confidence curves further verify that
when the confidence threshold is approximately 0.380, the model can achieve the
optimal balance between recall and precision (with an F1-score of 0.95 at this
point).


Figure 11: Confusion Matrix Diagram of Training Results
From the results of the confusion matrix, the model shows
good recognition performance for core colonies: in the normalized confusion
matrix, the correct classification ratio of these categories is close to or
reaches 1.0; in the original confusion matrix, the number of correctly
classified samples of core categories is large, and the number of misclassified
samples is small, indicating that the model can accurately and stably
distinguish core colonies.

Figure 12: Loss Curves and Evaluation Metric Curves
These curves indicate that during the training process,
various types of losses converge rapidly and eventually stabilize; core metrics
such as precision, recall, and mAP continue to rise and tend to saturate. This
shows that the model converges well on the colony detection task, being able to
accurately classify colony categories and regress bounding boxes, and has good
adaptability to different detection scenarios.
3.2.4 Practical Application
After completing the model training, we used the best
training weights obtained from the training to perform colony detection on the
images acquired from the experimental plating. Due to issues such as poor
detection results and small differences in colony states between adjacent time
points for some images, which provide no substantial help for subsequent data
analysis, we excluded these images from the candidate list. Finally, we
retained a batch of images with high detection accuracy that can clearly
reflect the dynamic growth process of colonies, and added a confidence filter
(confidence = 0.2). The detection results are as follows:

Figure 13: Diagram of Practical Application
(Specific colony information is displayed in the "Data
Analysis" module)
3.2.5 Discussion
From the results, although most colonies can be recognized
and the boundary recognition is relatively accurate, the following problems
still exist:
1. A small number of colonies cannot be detected;
2. Unclear large detection boxes appear in blank areas
without colonies;
3. Although the machine training effect diagram is good,
there are still cases of incorrect colony type recognition in practical
applications.
Based on the above problems and combined with the foundation
of our project, we speculate that the possible reasons are as follows:
① Since our experiment involves the growth of E.coli
from small to large, there are E.coli of various sizes in different
growth stages. Moreover, our dataset is not particularly large, which may make
it difficult to fully cover the morphologies of E.coli in all stages. At
the same time, the appearances of other types of colonies are similar to those
of E.coli in these stages, leading to machine misjudgment.
② At the same time, considering that some bacteria have
similar morphological characteristics themselves, and under the interference of
shooting methods or the environment, the core visual features of colony
morphologies are highly similar. Relying solely on the recognition method of
"image morphology" is prone to confusion.
③ In addition, during the shooting process, due to factors
such as light or reflection, some blank areas without colonies may present
"pseudo-features" similar to colonies, leading to machine
misjudgment. It may also be due to the Anchor-Free design adopted by YOLOv8s:
although it is suitable for targets of different sizes, the judgment threshold
for non-target areas is relatively low. When there are slight visual changes in
blank areas, such as background color gradients and tiny impurities, the model
tends to incorrectly calculate large-sized bounding boxes. If the subsequent
non-maximum suppression parameters are set too loosely to filter out these
false-positive large boxes, they may be retained in the detection results.
Combining the current machine training results and possible
existing problems, our future optimization directions will focus on targeted
dataset supplementation, experimental scenario expansion, algorithm
optimization, and so on.






 :
:
 to
to  .
.  can also be phosphorylated:
can also be phosphorylated:

 can efficiently phosphorylate FixJ to FixJ~P, while
can efficiently phosphorylate FixJ to FixJ~P, while  only performs inefficient phosphorylation:
only performs inefficient phosphorylation:
 acts as a potent phosphatase, and FixJ-P also undergoes spontaneous
dephosphorylation:
acts as a potent phosphatase, and FixJ-P also undergoes spontaneous
dephosphorylation: