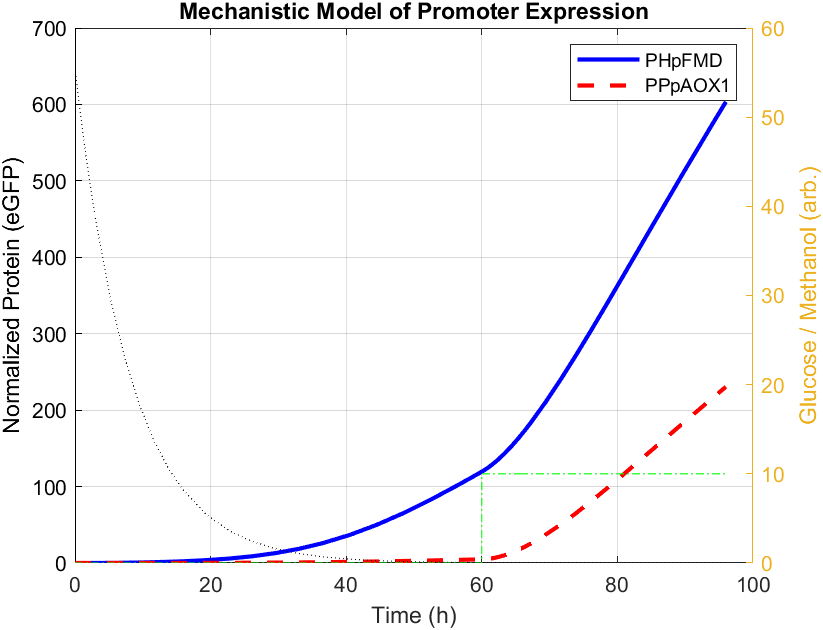
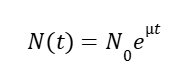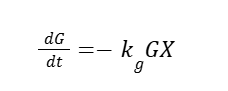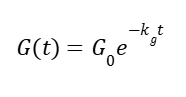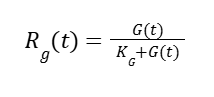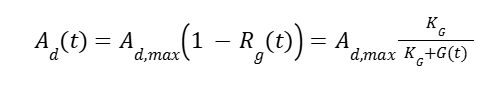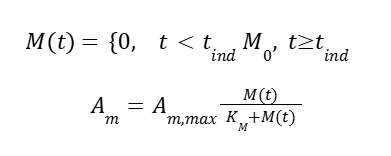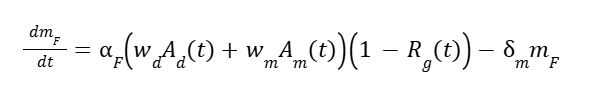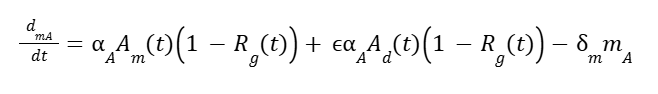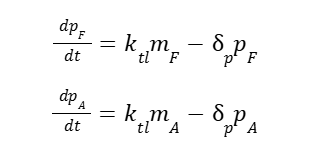We built a mechanistic ordinary differential equation (ODE) to represent the dynamics of both promoters HpFMD and AOX1. Both promoters are depressed by glucose, upon glucose depletion HpFMD shows strong derepressed activity, while AOX1 has very little expression until methanol is introduced. The formulation assumes that biomass is fixed. For glucose dynamics:
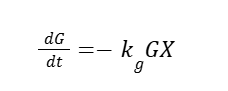
As glucose concentration (G, mM) is consumed in time t (h), where kg is the specific rate of consumption and X is biomass. Since we assume a normalize, steady biomass (X≈1):
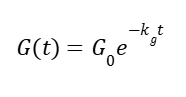
Where G0 is the initial concentration of glucose. Repressor activity is:
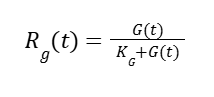
Where KG is the repression rate dependent on glucose. HpFMD has a strong de-repression activator in absence of glucose:
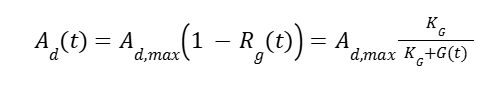
Where Ad is the de-repressor activator and Ad,max is the activator at maximum activity. Both are strongly induced under methanol added at induction time tind:
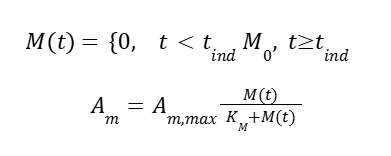
Where Am is the activity of the methanol induced activator, and KM is its specific rate. This leaves mRNA dynamics for HpFMD:
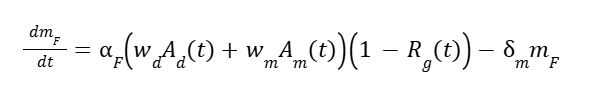
And for AOX1 is:
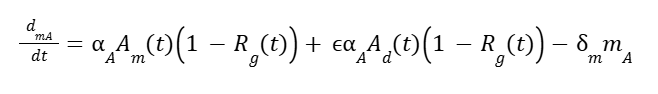
Where αF and αA are the max transcription rates, wd and wm are weights for de-repression vs methanol activation, ϵ is the weak de-repression factor for AOX1, m is mARN for AOX1 (A) and HpFMD (F), and δ is the degradation rate for mRNA
The protein dynamics for the same protein GFP (Vogl et al., 2020):
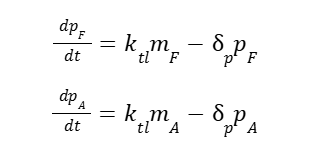
Where p is the protein production for AOX1 (A) and HpFMD (F).
We built both models in MATLAB R2023a.