Precision in design, excellence in execution.
Basic Parts
| Registry Code | Part Name | Function | Length (bp) |
|---|---|---|---|
| BBa_25J460FA⤴ | TMD | Encodes the transmembrane domain of the CD4 glycoprotein, enabling stable anchoring of fusion proteins to the plasma or vesicle membrane. | 77 |
| BBa_25Q8SCNI⤴ | Notchcore | Encodes the transmembrane core domain of the Notch receptor, mediating membrane integration and potential receptor-like topology for fusion constructs. | 978 |
| BBa_252HSMKK⤴ | CAAX membrane anchor | Encodes the CAAX prenylation motif, directing attached proteins to the inner leaflet of the plasma membrane via lipid modification. | 45 |
| BBa_25FPJ465⤴ | LifeAct | Encodes a 17–amino acid peptide that specifically binds filamentous actin (F-actin) without interfering with actin polymerization dynamics, enabling cytoskeletal localization of fusion proteins. | 51 |
| BBa_255Z07L1⤴ | KDEL | Encodes a C-terminal retention motif that retains soluble proteins in the endoplasmic reticulum through the KDEL receptor retrieval pathway. | 12 |
| BBa_25Q2M02J⤴ | Mitochondrial Targeting Sequence (MTS) | Encodes an N-terminal mitochondrial signal peptide that directs fusion proteins to the mitochondrial matrix or inner membrane. | 75 |
| BBa_250O67UT⤴ | Kozak sequence | Provides a consensus sequence surrounding the start codon to enhance ribosomal recognition and translation initiation efficiency in mammalian cells. | 10 |
| BBa_258ZYB5B⤴ | IgK–Nluc | Encodes a secreted form of NanoLuc luciferase driven by the Igκ signal peptide, serving as a luminescent reporter for secretion assays. | 597 |
| BBa_25WDV48R⤴ | mMaple3 | Encodes a photoconvertible fluorescent protein responsive to blue–violet light, used as a light-sensitive regulatory element or reporter. | 711 |
| BBa_25K5NJ9H⤴ | Furin Site | Encodes a protease recognition sequence cleaved by endogenous furin during vesicle trafficking, enabling post-Golgi release of cargo proteins. | 15 |
| BBa_25RHALHC⤴ | CarH | Encodes a green-light–responsive sensor protein that dissociates from oligomers into monomers upon illumination, allowing optogenetic control of protein–protein interactions. | 615 |
| BBa_25HYFBUH⤴ | NS3a | Encodes the NS3/4A serine protease domain that forms reversible complexes with Binder or GNCR proteins, functioning as a drug-responsive protein–protein interaction switch activated by Grazoprevir. | 588 |
| BBa_25CQAUVL⤴ | Apo NS3a leader (Binder) | Encodes a high-affinity protein partner that binds NS3a in the absence of Grazoprevir and dissociates rapidly upon drug treatment, enabling small-molecule–controlled complex disruption. | 63 |
| BBa_25DRNV7I⤴ | GNCR | Encodes a high-affinity protein interacting with NS3a in a Grazoprevir-dependent manner, serving as an alternative small-molecule–responsive binding partner. | 699 |
Favourite Basic Part
NS3a(BBa_25HYFBUH)
Overview
The hepatitis C virus (HCV) NS3/4A protease (NS3a) is a critical enzyme for viral replication and a well-established drug target. Its activity can be competitively inhibited by compounds such as the small-molecule drug Grazoprevir1 and the short-peptide Binder, which bind to its active site.
Beyond its biological role, NS3a has emerged as a foundational component in synthetic biology due to these specific, drug-regulatable interactions. Its core utility lies in a dual-control mechanism: it naturally forms a high-affinity (nanomolar) complex with Binder, which Grazoprevir can efficiently dissociate2. Concurrently, the same drug acts as a molecular bridge, enabling NS3a to recruit a computationally designed protein, GNCR3. This unique capacity for both chemically induced dissociation (CIDiss) from Binder and chemically induced dimerization (CIDimer) with GNCR makes NS3a an exceptionally versatile platform for engineering sophisticated genetic switches.

Figure 1. Schematic illustration of the mechanism of Grazoprevir induced NS3a-Binder dissociation or NS3a-GNCR interaction
Experimental Validation
Methods
Cell Culture and Transfection
HEK293T cells (ATCC CRL-3216) were cultured in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin at 37 °C and 5% CO₂. Cells were seeded at 5 × 104 per well in 24-well plates and transfected 12 h later using a PEI-based protocol (PEI:DNA = 5:1, w/w). The transfection mix was replaced with fresh medium 6 h post-transfection. Unless indicated otherwise, transfection was performed at 16 h after seeding 5 × 104 mammalian cells into each well of a 24-well plate. The cell culture medium was replaced with fresh medium (not containing transfection reagents) at 6 h after transfection. HEK-293T cells were transfected using a PEI-based protocol at a PEI:DNA ratio of 5:1 (w/w) and in a transfection volume of 50 μL native serum-free DMEM per well.
Reporter Quantification
Secreted NanoLuc levels were measured from cell culture supernatants using the Nano-Glo Luciferase Assay System (Promega, N1120). All measurements were performed in biological quadruplicates (n = 4). Statistical analyses used two-tailed unpaired t-tests (P < 0.05 considered significant)
Characterization
NS3a's function in CIDiss switch configurations
For the CIDiss switch based on Grazoprevir-induced NS3a-Binder dissociation, HEK cells were co-transfected with three plasmids: PCMV-IgK-Fluc-TMD-NS3a-BGHPA (BBa_2506II6Z), PCMV-Binder-TetR-NLS-VP64-BGHPA (BBa_2596NLG0), and TCE-IgK-Nluc-BGHPA (BBa_25EPQC3P) at a 1:2:6 DNA ratio (corresponding to 50 ng, 100 ng, and 300 ng per well, respectively). At 24 hours post-transfection, cells were treated with 10 µM Grazoprevir (experimental group) or an equivalent volume of DMSO (vehicle control). Supernatants were harvested 24 hours after drug treatment for NanoLuc luciferase quantification. The results showed a significant increase in NanoLuc activity in Grazoprevir-treated cells compared to the control, confirming the successful establishment of a functional NS3a-based CIDiss system capable of small molecule-regulated gene expression.
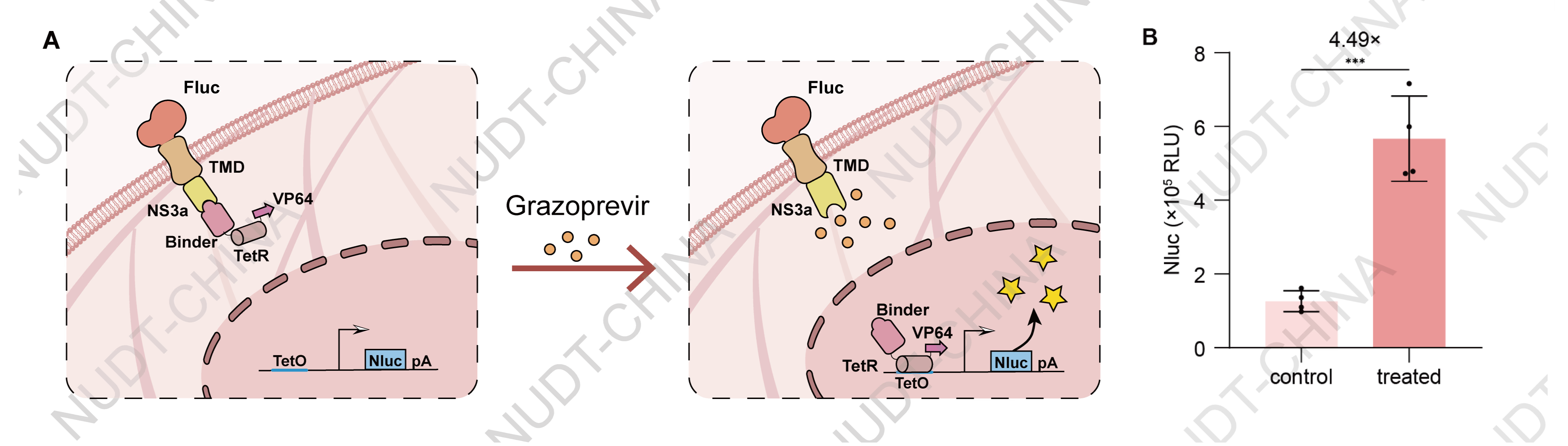
Figure 2. NS3a's function in CIDiss switch configurations. NanoLuc levels in culture supernatants 24 hours after Grazoprevir (10 µM) or DMSO treatment. Data shown as mean ± SD (n = 4); ***P < 0.001.
NS3a's function in CIDimer switch configurations
For the CIDimer switch based on Grazoprevir-induced NS3a:GNCR interaction, HEK cells were co-transfected with three plasmids: PCMV-TetR-NLS-NS3a-BGHPA (BBa_25NDCL0K), PCMV-GNCR-GSlinker-NLS-VP64-BGHPA (BBa_25PW9O16), and TCE-IgK-Nluc-BGHPA (BBa_25EPQC3P) at a 2:2:1 DNA ratio (corresponding to 200 ng, 200 ng, and 100 ng per well, respectively). At 24 hours post-transfection, cells were treated with 10 µM Grazoprevir (experimental group) or an equivalent volume of DMSO (vehicle control). Supernatants were harvested 24 hours after drug treatment for NanoLuc luciferase quantification. The results showed a significant increase in NanoLuc activity in Grazoprevir-treated cells compared to the control, confirming the successful establishment of a functional NS3a-based CIDimer system capable of small molecule-regulated gene expression.
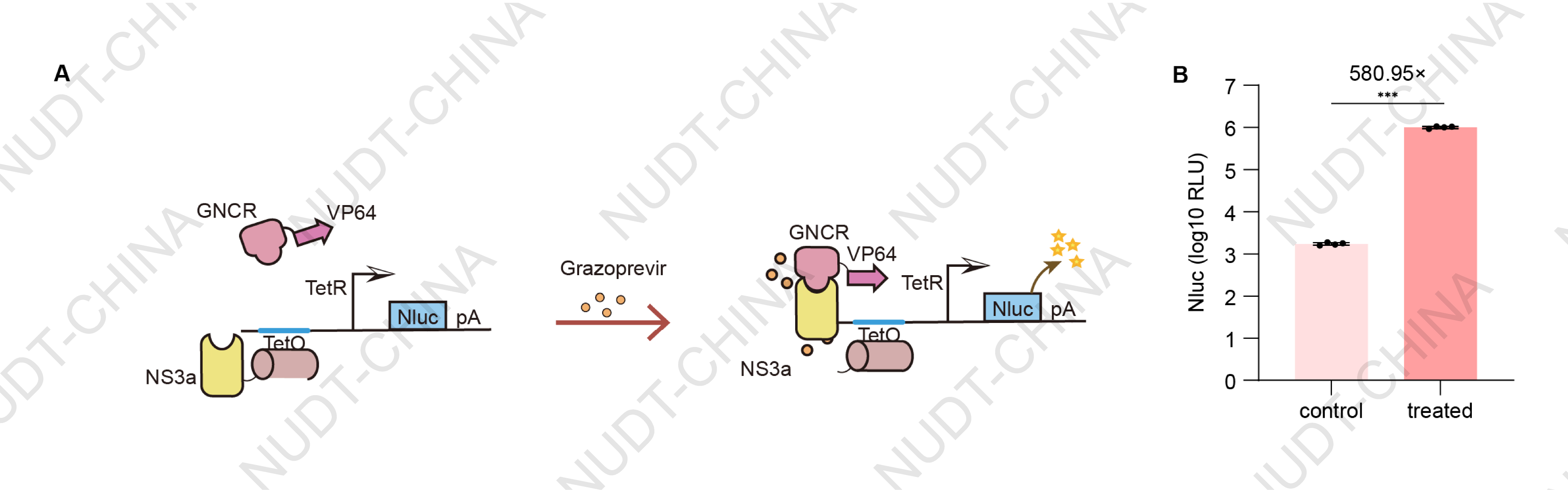
Figure 3. NS3a's function in CIDimer switch configurations. NanoLuc levels in culture supernatants 24 hours after Grazoprevir (10 µM) or DMSO treatment. Data shown as mean ± SD (n = 4); ***P < 0.001
Application
In our project, NS3a (BBa_25HYFBUH) was directly incorporated into the core response module and collaborated with the anchoring module to jointly construct a modular secretion control system(chemSPARK ,Figure 4A). By leveraging the Grazoprevir-induced NS3a-Binder dissociation mechanism, we successfully developed a molecular switch capable of rapidly responding to drug signals and precisely controlling the secretion of target proteins. This fully demonstrates the powerful functionality and application potential of this foundational component within complex genetic circuits.
HEK293T cells were co-transfected with plasmids carrying PCMV–IgK–(Nluc–FS)3–TMD–(NS3a)2–BGHPA (BBa_25WEPAKX) and plasmids carrying PCMV-Binder–mCherry–LifeAct–BGHPA (BBa_25NYLBFF) at a 1:3 DNA ratio (100 ng : 300 ng per well).After 48 h, cells were treated with 10 µM Grazoprevir (experimental group) or DMSO (control). Supernatants were collected 2 h post-treatment for NanoLuc quantification. Cells treated with Grazoprevir showed a significant increase in NanoLuc secretion, confirming that NS3a–Binder dissociation releases pre-anchored vesicles and activates rapid protein secretion (Figure 4B).
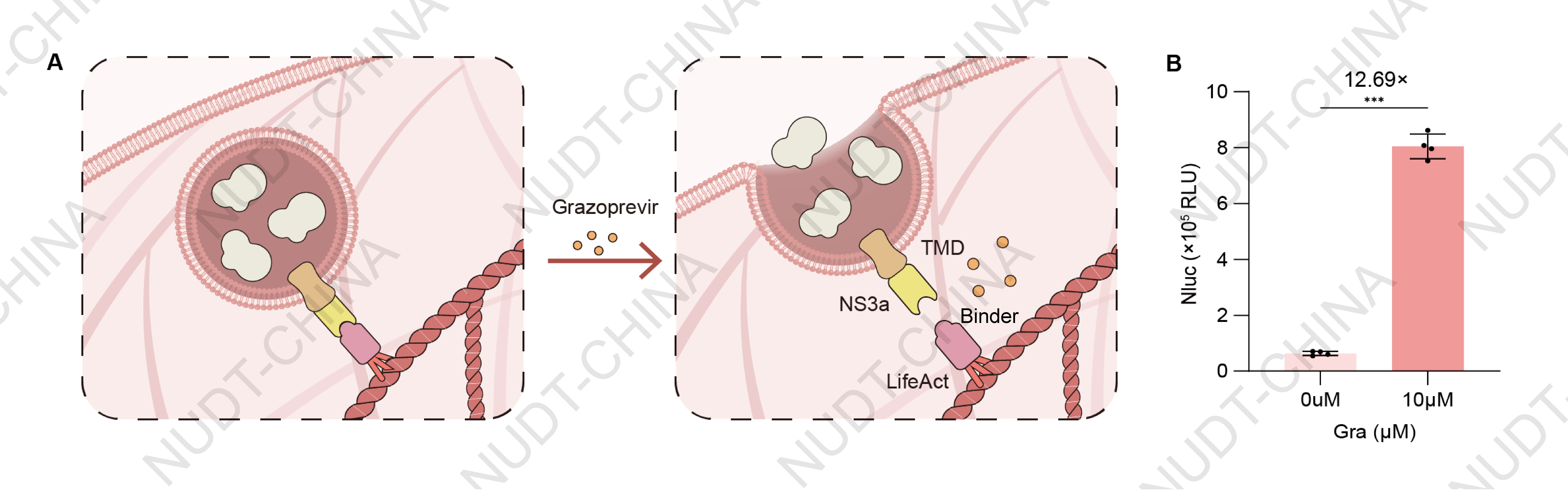
Figure 4. NanoLuc levels in culture supernatants 2 h after Grazoprevir (10 µM) or DMSO treatment. Data shown as mean ± SD (n = 4); ***P < 0.001.
Discussion
Limitations:
In both transcriptional and secretory regulation contexts, the NS3a-based system functions as a drug-inducible module whose activity depends on the continued presence of its small-molecule inducer. Prolonged activation may be affected by drug metabolism, proteolytic degradation of the NS3a complex, or potential cytotoxic and off-target effects of the compound. These factors must be carefully balanced when designing long-term or high-sensitivity applications.
Comparisons:
Compared with optogenetic systems such as mMaple3 and CarH, which achieve noninvasive and precise light-based control, the NS3a-based system trades optical precision for pharmacological convenience. By relying on a diffusible small molecule rather than light, it overcomes limitations in tissue penetration and experimental setup, enabling stable and systemic activation suitable for in vivo or clinical scenarios that demand long-duration control.
Future Directions:
A key advantage of the NS3a platform is its use of clinically approved drugs such as Grazoprevir, which confers excellent biocompatibility and translational potential. This feature positions the NS3a-based switch as a valuable tool for drug-responsive disease modeling and high-throughput screening in mammalian cells. Beyond research use, it offers a foundation for drug-controllable therapeutic systems, allowing precise regulation of gene expression or secretion in engineered cells. Whether applied to long-term transcriptional modulation or rapid post-translational secretion, NS3a-based regulation provides a flexible and clinically relevant strategy for next-generation smart cell therapies.
 Description
Description
 Design
Design
 Notebook
Notebook
 Results
Results
 Basic Parts
Basic Parts
 Composite Parts
Composite Parts
 Parts Collection
Parts Collection
 Education
Education
 Art Gallery
Art Gallery
 Implementation
Implementation
 Attributions
Attributions
 Collaboration
Collaboration