Subcellular Localization Part Collection: A Modular Toolkit for Spatial Control in Mammalian Synthetic Biology
Overview
Subcellular localization elements are fundamental design modules in mammalian synthetic biology. By directing proteins to specific intracellular compartments, such as the plasma membrane, mitochondria, nucleus, or cytoskeleton, engineers can precisely control enzymatic reactions, signal transduction, and secretion at the spatial level.
Our Subcellular Localization Toolkit compiles a coherent set of targeting elements that function as interchangeable modules for positioning proteins within cells. These parts support a wide range of applications, including:
- Compartmentalized metabolic engineering and toxin isolation,
- Enhanced protein folding and post-translational modification,
- Light- or drug-controlled nuclear transport,
- Programmable secretion and signal activation.
Together, these modules provide a standardized and expandable platform for spatial control of biological functions, improving the precision, safety, and tunability of mammalian synthetic biology systems.
Parts in this collection
This toolkit is a coordinated collection of cellular localization tags, which were experimentally validated through live-cell confocal imaging in HeLa or HEK293T cells, demonstrating distinct localization patterns that matched their designed cellular destinations.
| Localization | Representative Parts | Registry IDs | Function | Length (bp) |
|---|---|---|---|---|
| Secretion signal Peptide | IgK signal | BBa-K4646003⤴ | Directs proteins into the secretory pathway | 45 |
| Cell membrane localization | TMD, NotchCore, CAAX | BBa_25J460FA⤴, BBa_25Q8SCNI⤴, BBa_252HSMKK⤴ | Targets proteins to the plasma membrane for controlled secretion | 77, 978, 45 |
| Mitochondrial localization | MTS (Mitochondrial Targeting Sequence) | BBa_25Q2M02J⤴ | Directs proteins to mitochondria for energy-related processes | 75 |
| Nuclear import/export | NLS, NES | BBa_K404153⤴, BBa_K4414003⤴ | Mediates nuclear entry or export for transcriptional regulation | 21, 30 |
| Cytoskeletal localization | LifeAct | BBa_25FPJ465⤴ | Anchors proteins to F-actin for spatial and dynamic control | 51 |
Documentation and Standardization
Documentation and Standardization
All modules were experimentally validated and comprehensively documented in the iGEM Registry(see our collection page: Subcellular Localization System). Each part entry includes full sequences, construct schematics, characterization methods, and high-resolution confocal data collected under standardized imaging conditions.
Experimental Characterization
Cell membrane localization: TMD, NotchCore, and CAAX modules expressed in HeLa cells co-localized with plasma membrane dyes, confirming precise membrane targeting.
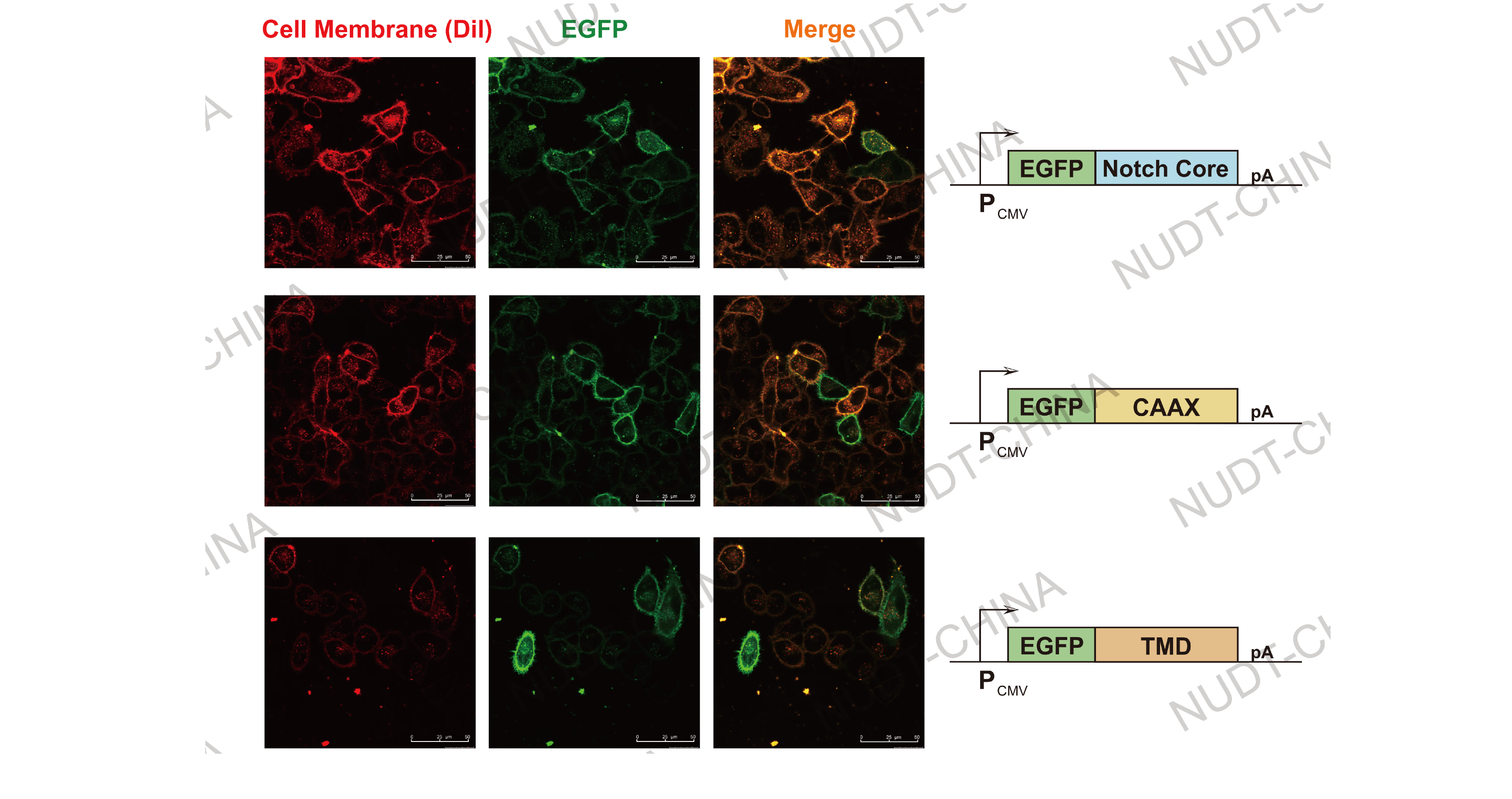
Figure 1. Confocal micrographs of membrane-localized proteins.
Cell Membrane (Red, stained with Dil stain) and EGFP fluorescent (Green) are shown. Images were taken 24 hours post transfection. Scale bars, 50μm
Mitochondrial localization: MTS-tagged constructs showed punctate or tubular fluorescence consistent with mitochondrial networks.
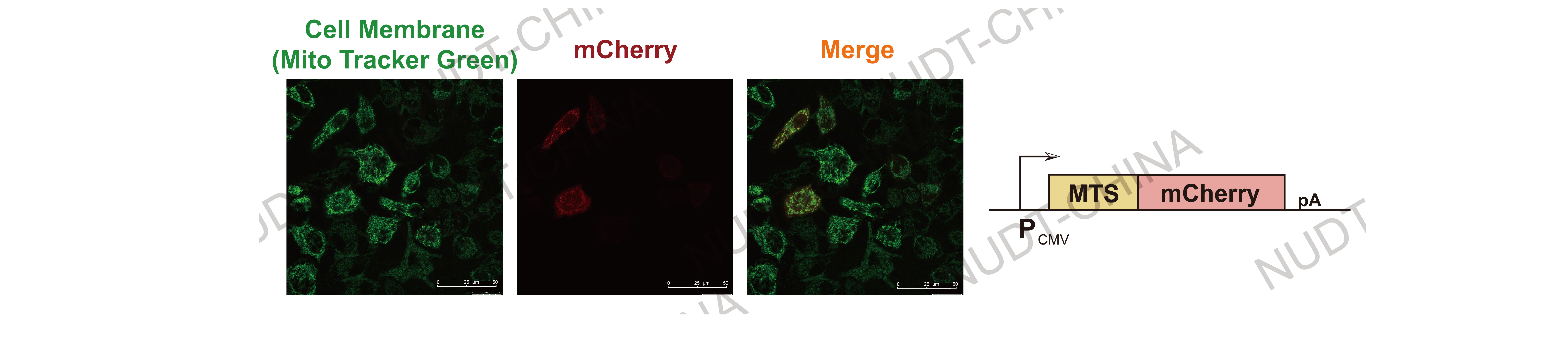
Figure 2. Confocal micrographs of mitochondrial localization.
Mitochondria (Green, stained with Mito Tracker Green) and mCherry fluorescent (Red) are shown. Images were taken 24 hours post transfection Scale bars, 50μm
Nuclear localization and export: NLS and NES tags generated distinct nuclear enrichment or exclusion of GFP reporters, verifying direction-specific transport.
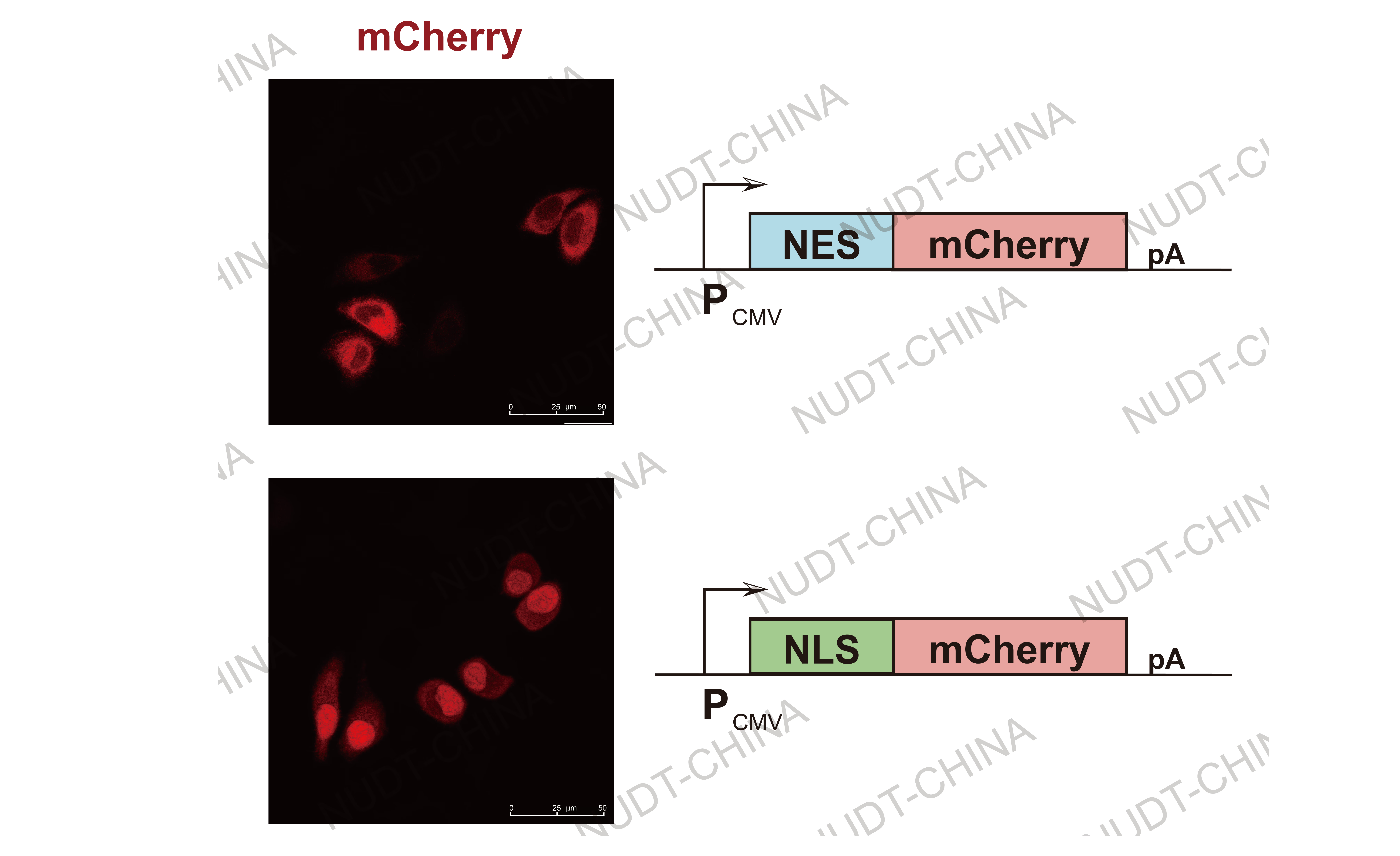
Figure 3. Confocal micrographs of nuclear localization/export.
NES-mChery (Upper panel) and NLS-mCherry (Lower panel) are shown. Images were taken 24 hours post transfection Scale bars, 50μm
Cytoskeletal localization (LifeAct): LifeAct-tagged proteins displayed filamentous fluorescence aligned with actin filaments, demonstrating accurate cytoskeletal anchoring. LifeAct also served as the core anchoring module for our SPARK secretion system.
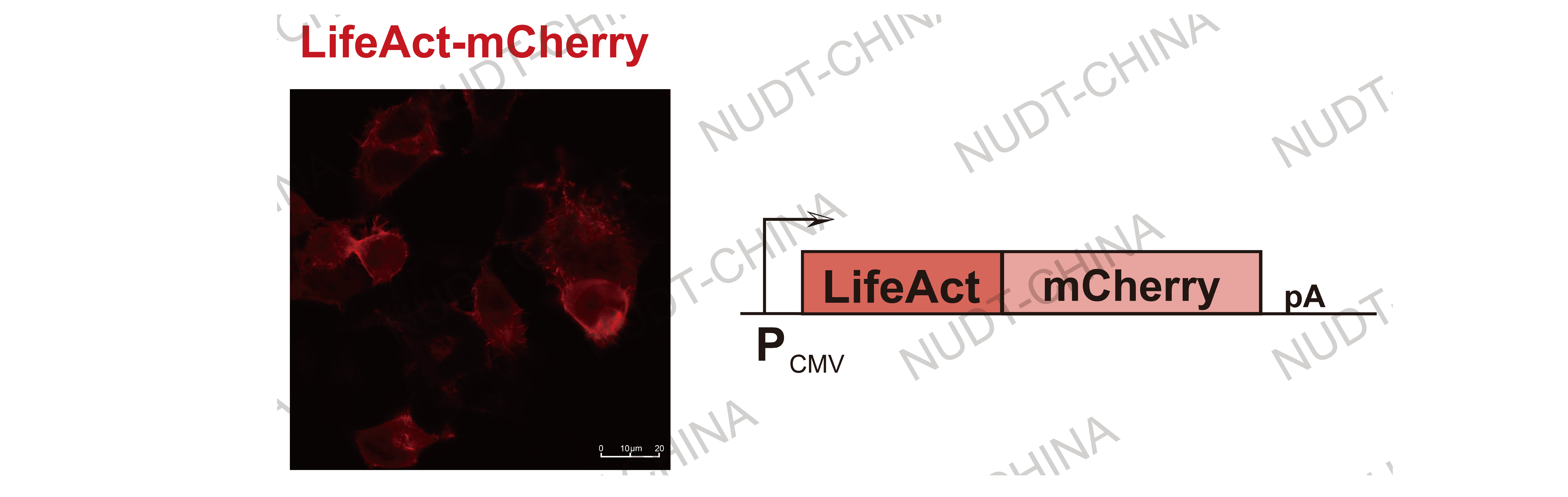
Figure 4. Confocal micrographs of cytoskeletal localization using LifeAct.
The LifeAct-mCherry (Red) exhibits the hole cytoskeleton. Images were taken 24 hours post transfection Scale bars, 10μm
Secretion signal: Fusion of the IgK signal sequence to NanoLuc reporters resulted in efficient extracellular secretion, confirming proper trafficking through the ER--Golgi pathway.
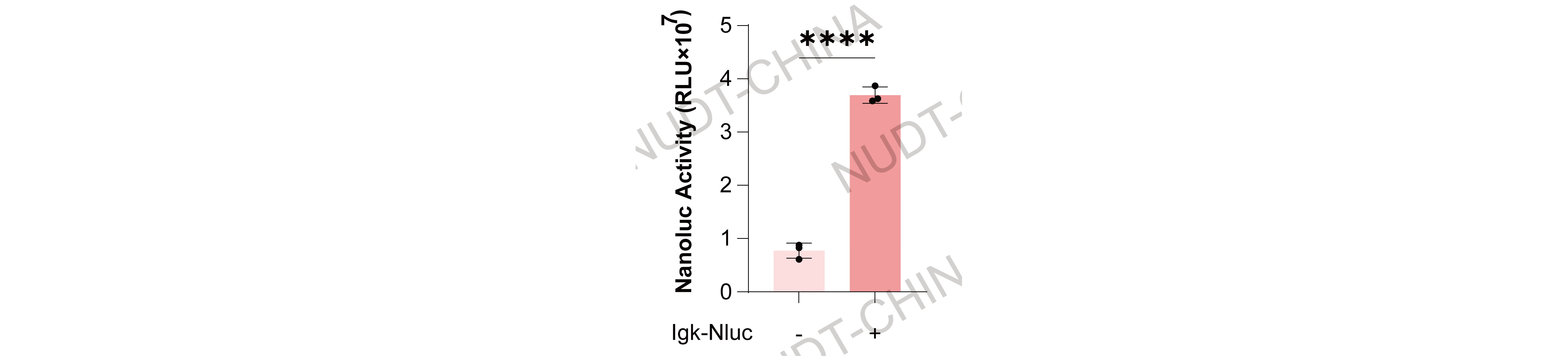
Figure 5. NanoLuc levels in culture supernatants 24 h after change medium. Data shown as mean ± SD (n = 3); ***P < 0.001.
Documentation and Standardization
All modules were experimentally validated and comprehensively documented in the iGEM Registry(see our collection page: Subcellular Localization System). Each part entry includes full sequences, construct schematics, characterization methods, and high-resolution confocal data collected under standardized imaging conditions.
Experimental Characterization
Cell membrane localization: TMD, NotchCore, and CAAX modules expressed in HeLa cells co-localized with plasma membrane dyes, confirming precise membrane targeting.

Figure 1. Confocal micrographs of membrane-localized proteins.
Cell Membrane (Red, stained with Dil stain) and EGFP fluorescent (Green) are shown. Images were taken 24 hours post transfection. Scale bars, 50μm
Mitochondrial localization: MTS-tagged constructs showed punctate or tubular fluorescence consistent with mitochondrial networks.

Figure 2. Confocal micrographs of mitochondrial localization.
Mitochondria (Green, stained with Mito Tracker Green) and mCherry fluorescent (Red) are shown. Images were taken 24 hours post transfection Scale bars, 50μm
Nuclear localization and export: NLS and NES tags generated distinct nuclear enrichment or exclusion of GFP reporters, verifying direction-specific transport.

Figure 3. Confocal micrographs of nuclear localization/export.
NES-mChery (Upper panel) and NLS-mCherry (Lower panel) are shown. Images were taken 24 hours post transfection Scale bars, 50μm
Cytoskeletal localization (LifeAct): LifeAct-tagged proteins displayed filamentous fluorescence aligned with actin filaments, demonstrating accurate cytoskeletal anchoring. LifeAct also served as the core anchoring module for our SPARK secretion system.

Figure 4. Confocal micrographs of cytoskeletal localization using LifeAct.
The LifeAct-mCherry (Red) exhibits the hole cytoskeleton. Images were taken 24 hours post transfection Scale bars, 10μm
Secretion signal: Fusion of the IgK signal sequence to NanoLuc reporters resulted in efficient extracellular secretion, confirming proper trafficking through the ER--Golgi pathway.

Figure 5. NanoLuc levels in culture supernatants 24 h after change medium. Data shown as mean ± SD (n = 3); ***P < 0.001.
Functional Integration and Community Utility
The collection was used to build a fully functional post-translational secretion control system (SPARK, see our Engineering and Results Page), in which LifeAct mediated vesicle anchoring and release upon signal induction, secretion peptide and TMDs mediated the integration of POI into the secretion vesicles. This integration verifies that the toolkit functions not only as individual parts but also as a modular system enabling complex cellular behaviors.
Beyond SPARK, the toolkit supports:
- Custom intracellular protein positioning for pathway engineering,
- Orthogonal compartmentalization for multi-enzyme cascades,
- Rapid prototyping of spatially resolved genetic circuits.
All modules conform to BioBrick standards and are optimized for mammalian expression, making them immediately accessible and reusable for future iGEM teams and researchers.
The collection was used to build a fully functional post-translational secretion control system (SPARK, see our Engineering and Results Page), in which LifeAct mediated vesicle anchoring and release upon signal induction, secretion peptide and TMDs mediated the integration of POI into the secretion vesicles. This integration verifies that the toolkit functions not only as individual parts but also as a modular system enabling complex cellular behaviors.
Beyond SPARK, the toolkit supports:
- Custom intracellular protein positioning for pathway engineering,
- Orthogonal compartmentalization for multi-enzyme cascades,
- Rapid prototyping of spatially resolved genetic circuits.
All modules conform to BioBrick standards and are optimized for mammalian expression, making them immediately accessible and reusable for future iGEM teams and researchers.
Conclusion
The Subcellular Localization Toolkit provides a coherent, experimentally verified, and community-oriented collection that provides:
- A unified and modular design framework,
- Comprehensive documentation and validation consistent with Registry standards,
- A functional system (SPARK) built using the collection,
- Broad practical value for advancing mammalian synthetic biology.
The Subcellular Localization Toolkit provides a coherent, experimentally verified, and community-oriented collection that provides:
- A unified and modular design framework,
- Comprehensive documentation and validation consistent with Registry standards,
- A functional system (SPARK) built using the collection,
- Broad practical value for advancing mammalian synthetic biology.
 Description
Description
 Design
Design
 Notebook
Notebook
 Results
Results
 Basic Parts
Basic Parts
 Composite Parts
Composite Parts
 Parts Collection
Parts Collection
 Education
Education
 Art Gallery
Art Gallery
 Implementation
Implementation
 Attributions
Attributions
 Collaboration
Collaboration