
For astaxanthin biosynthesis in E.coli, seven key genes including HpCrtE, PanCrtB, PanCrtI, PagCrtY, idi, HpCrtZ, BreCrtW, were chemically synthesized by GenScript Biotech Corporation (Nanjing, China) and inserted into the commonly used bacteria vector pET-Duet-1 for expression as following design (Fig. 1).
Briefly, CDs of HpCrtE, PanCrtI, and PanCrtB were sequentially followed by the first T7 promoter while CDs of PagCrtY, idi, HpCrtZ, BreCrtW were sequentially followed by the second T7 promoter. The 14 bp nucleotide was used to space each gene. The synthesized plasmid was delivered into E.coli Top10 cells for storage.
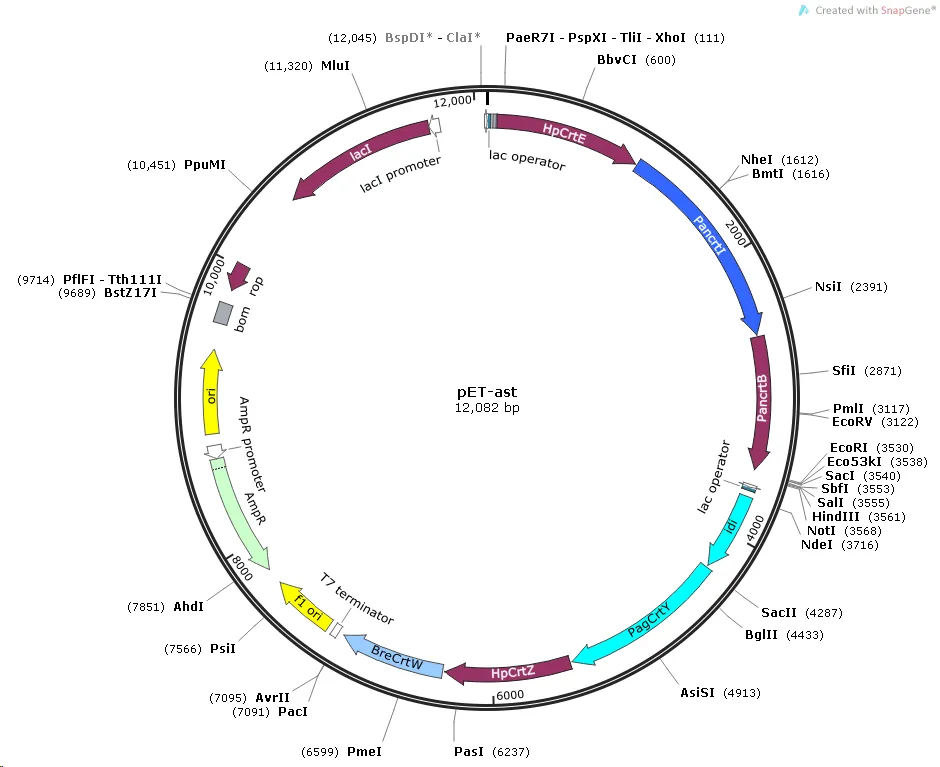 Figure 1. The schematic map of plasmid used for astaxanthin production in E. coli .
Figure 1. The schematic map of plasmid used for astaxanthin production in E. coli .
Plasmid was extracted from E. coli cells using Omega Plasmid Mini Kit. Detailed procedures are as following:
The obtained plasmid DNA was quantified by NanoDrop 2000 and stored at -20°C for future use.
Prepare the reaction in a 0.2 mL EP tube on ice as shown in Table 1. After adding all agents, briefly centrifuge the mixture and then incubate at 37°C for 1 hour.
| Agent | Amount |
|---|---|
| Plasmid DNA | 1 µg |
| FastDigest Pac I | 1 µL |
| FastDigest Nde I | 1 µL |
| 10 × FastDigest Green Buffer | 2 µL |
| ddH₂O | To final 20 µL |
To prepare 1% agarose gel, detailed procedures are as following:
E. coli BL21(DE3) was used as the factory for pigment production. Competent cells of BL21(DE3) were purchased from Weidi Biotech (Weidi Biotechnology Co., Ltd, Shanghai, China). Heat-shock method was used for E. coli transformation. Detailed procedures are as following:
To prepare 1 L LB medium, 10 g of sodium chloride, 10 g of peptone, and 5 g of yeast extract were completely dissolved in dH₂O to final volume of 1 L. For solid medium preparation, additional 15 g of agar powder was added. The prepared medium was autoclaved at 121°C for 20 minutes.
Detailed procedures for seedling culture are as following:
In a laminar flow hood, 1 mL seed culture was inoculated into a 250 mL flask containing 100 mL LB medium supplied with 100 mg/ml ampicillin. The flask was put into a shaker at 30°C with 220 rpm for about 2 hours until OD₆₀₀ reaches 0.5-0.6. IPTG at a final concentration of 0.1 mM was added into the culture and cells were grown for another 5 hours.
Cells were harvested using 50 mL tube by centrifugation at 8000 rpm for 8 minutes at 4°C. To remove the residual culture medium, cells were washed by sterilized ddH₂O and harvested again by centrifugation at 8000 rpm for 8 minutes at 4°C[1].
Firstly, picked single colony containing the astaxanthin synthesis plasmid from the plates and cultured in 250 mL Erlenmeyer flasks containing 80 mL LB medium (100 μg/mL ampicillin). In a laminar flow hood, 80 mL seed culture was inoculated into a 1000 mL flask containing 800 mL LB medium supplied with 100 μg/mL ampicillin. The flask was put into a shaker at 30°C with 220 rpm until OD₆₀₀ reaches 0.6. IPTG at a final concentration of 0.1 mM was added into the culture and cells were grown for another 5 hours. Cells were harvested by centrifugation at 8000 rpm for 8 minutes at 4°C. To remove the residual culture medium, cells were washed by sterilized ddH₂O and harvested again by centrifugation at 8000 rpm for 8 minutes at 4°C[1].
Harvested cells were dried in a vacuum freeze dryer. Firstly, seal the centrifuge tube with cell pellet with parafilm and punch 5-10 small holes with toothpick. Secondly, freeze the cell pellet by liquid nitrogen. Lastly, place the frozen samples in a rack upstraight and put the rack into the cold trap chamber. After 48 hours, cell pellets were dried and subjected to pigment extraction.
Detailed procedures for pigment extraction are as following:
The astaxanthin content in the pigment extracts from E. coli cells was determined by HPLC. Shimadzu LC-2030C 3D system (Shimadzu, Kyoto, Japan) HPLC instrument coupled with Phenomenex Gemini-NX C18 column (5 µm, 150 × 3.0 mm, Phenomenex Inc., Aschaffenburg, Germany) was used.
The mobile phases include ultrapure water (Solution A) and methanol: isopropanol (8:2) (Solution B). The flow rate is 0.6 mL/min, injection volume was 10 µL, column temperature was 35 °C, and DAD detection at 450 and 475 nm.
The gradient for HPLC analysis was initial conditions at 15% A, 85% B; 0-10 min from initial condition to 100% B; 10-12 min maintained at 100% B; 12-12.1 min back to initial conditions; and 12.1-18 min recalibrate the column with initial condition. Chromatograms and the peak area of astaxanthin was recorded using LabSolution software.
Calibration curve was used to calculate the astaxanthin concentration in the detected sample. Firstly, commercial astaxanthin standards were prepared at the concentrations of 1, 0.5, 0.25, and 0.1 µg/mL. Subsequently, standards were run by HPLC. Finally, the concentrations of standards (y-axis) were plotted with corresponding peak areas (x-axis) to construct the standard curve.
The formula used for astaxanthin content calculation was:
Astaxanthin content in µg/mg DCW (dried cell weight) = asta (conc. µg/mL) × 1 (mL) / W (mg)
Where asta (conc. µg/mL) is the calculated astaxanthin concentration in the detected sample using peak area based on the standard curve and W is the exact weight of cells subjected for pigment extraction[2].
[1] Ma T, Zhou Y, Li X, Zhu F, Cheng Y, Liu Y, Deng Z, Liu T. Genome mining of astaxanthin biosynthetic genes from Sphingomonas sp. ATCC 55669 for heterologous overproduction in E. coli. Biotechnol J. 2016;11:228–237.
[2] Huang D, Liu W, Li A, Wang C, Hu Z. Discovery of geranylgeranyl pyrophosphate synthase (GGPPS) paralogs from Haematococcus pluvialis based on Iso-Seq analysis and their function on astaxanthin biosynthesis. Mar Drugs. 2019;17:696.