
The astaxanthin biosynthesis is a set of enzymatic reaction. To computationally predict our design on astaxanthin productivity improvement, models are constructed which are also helpful for better understanding the interaction between enzyme and substrate in biology. The targets in our wet lab were selected according to model analysis. Each model was listed below.
β-carotene hydroxylase from Haematococcus pluvialis (HpCrtZ) is a key enzyme which converted canthaxanthin to astaxanthan [1].We register it as a new element (BBa_25ESIC8B). The amino acid sequence of β-carotene hydroxylase was shown as following:
HpCrtZ:
MLSKLQSISVKARRAELARDITRPKVCLHAQRCSLVRLRVAAPQTEEAVGTVQAAGAGDEHSADVALQQLDRAIAERRARRKREQLSYQAAAIAASIGVSGIAIFATYLRFAMHMTVGGAVPWGEVAGTLLLVVGGALGMEMYARYAHKAIWHESPLGWLLHKSHHTPRTGPFEANDLFAIINGLPAMLLCTFGFWLPNVLGAACFGAGLGITLYGMAYMFVHDGLVHRRFPTGPIAGLPYMKRLTVAHQLHHSGKYGGAPWGMFLGPQELQHIPGAAEEVERLVLELDWSKR*
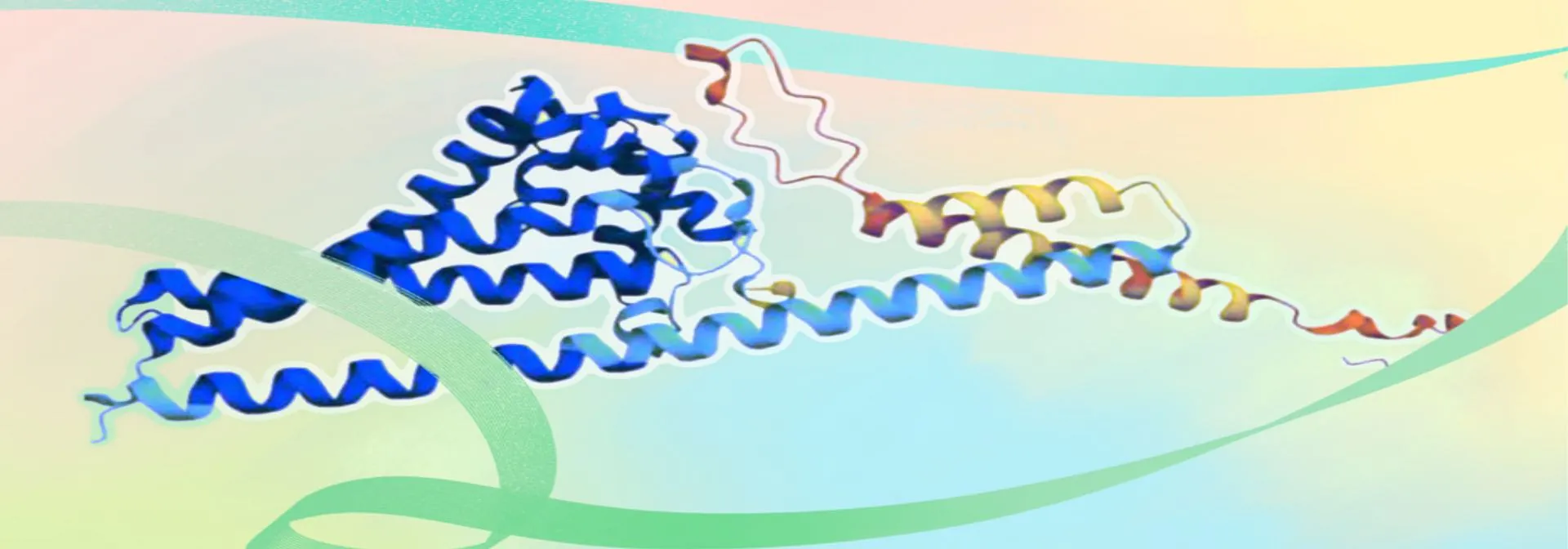
The structure file of β-carotene hydroxylase was obtained from the AlphaFold website under the accession Q9PK6 [2].It was imported into the Prankweb platform to generate 3D structural model. The β-carotene hydroxylase exhibits a highly helical conformation dominated by α-helices with a distinct hydrophobic pocket at the center (Figure 1).
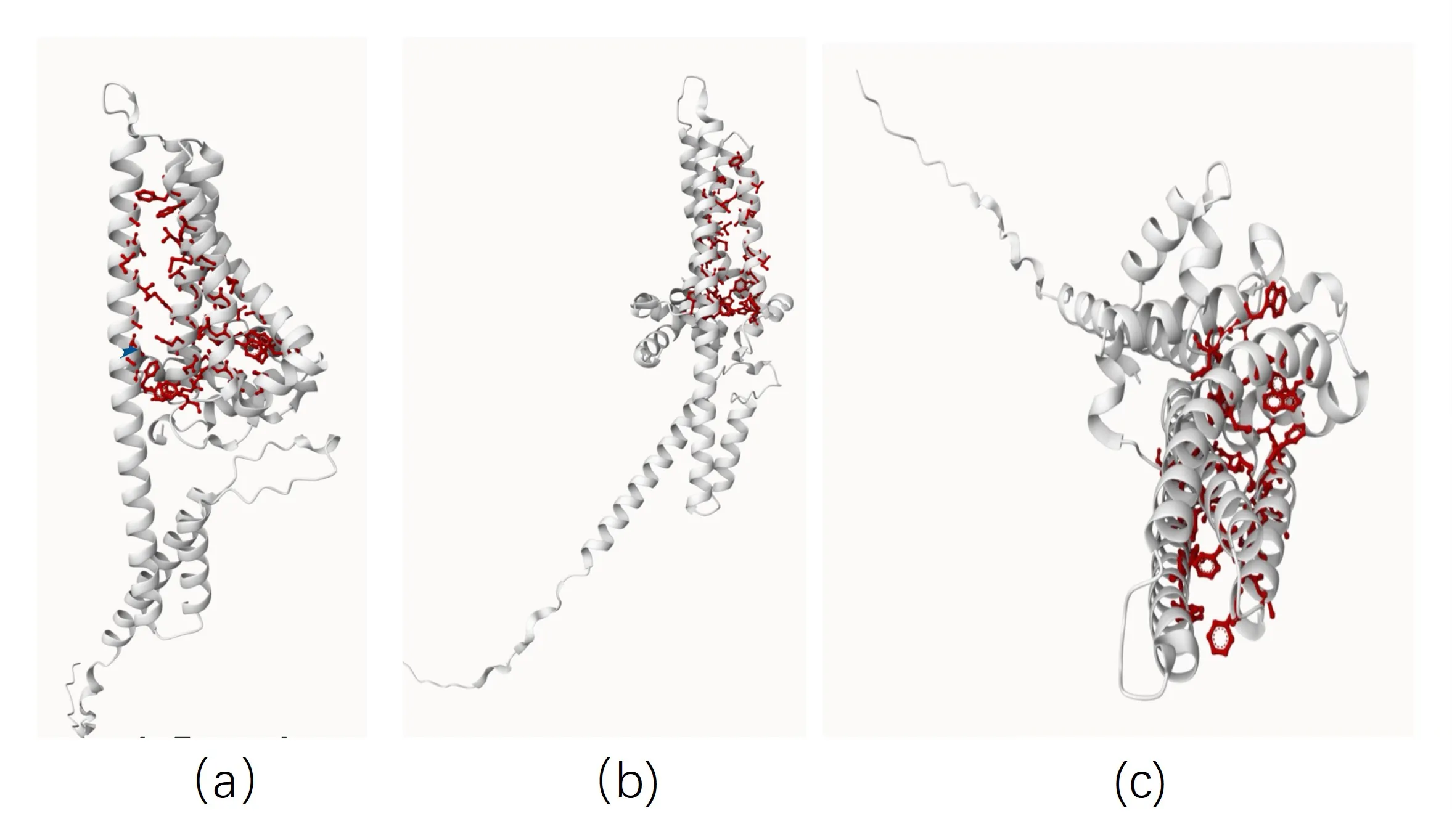 Figure 1. The 3D structure of β-carotene hydroxylase in front view (a), side view (b), and vertical view (c)
Figure 1. The 3D structure of β-carotene hydroxylase in front view (a), side view (b), and vertical view (c)
We obtained canthaxanthin and β-carotene from the PubChem database with CID 5281227 and CID 528048, respectively. Model files for β-carotene hydroxylase and substrates were uploaded to the CB-Dock2 website where molecular docking simulations were performed [3]. The binding sites of β-carotene hydroxylase with canthaxanthin were predicted to position at TYR88, TRP152, HIS165, GLU174, ASN176, ASP177, PHE179, ALA180, ASN183, GLY216, TYR219, MET220, HIS223, ASP224, ARG230(Figure 2). The binding sites of β-carotene hydroxylase with β-carotene were predicted to position at TYR88, VAL99, ILE102, ALA103, PHE105, ALA106, LEU109, MET140, TRP152, ASN176, ASP177, PHE179, ALA180, ASN183, GLY184, ALA187, MET188, CYS191, THR192, PHE195, TRP196, LEU210, ILE212, THR213, GLY216, MET217, TYR219, MET220, HIS223, ASP224(Figure 2b).
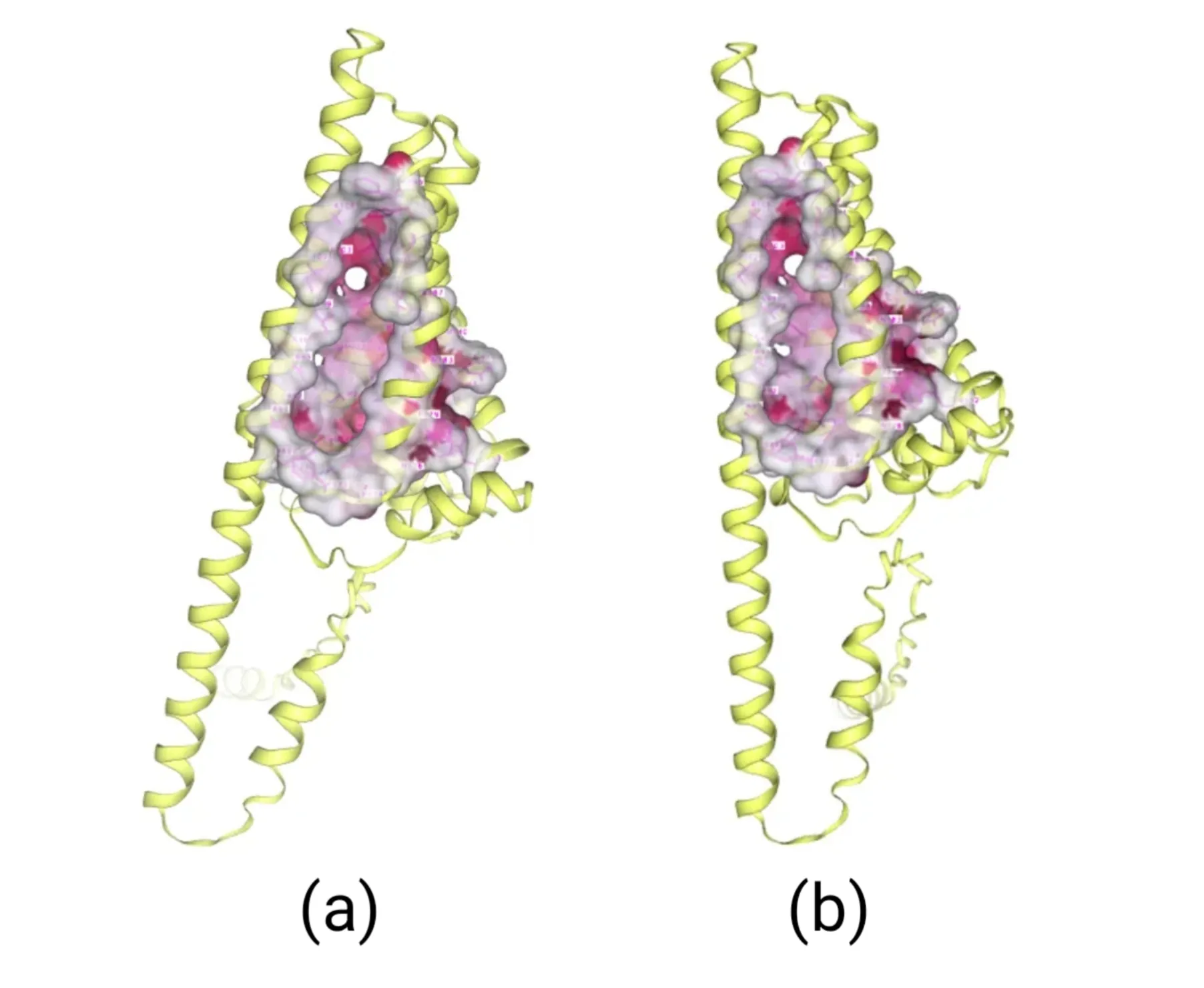 Figure 2. Predicted molecular docking between β-carotene hydroxylase and canthaxanthin (a) and β-carotene (b)
Figure 2. Predicted molecular docking between β-carotene hydroxylase and canthaxanthin (a) and β-carotene (b)
The pocket region constitutes the active site of β-carotene hydroxylase, and its substrate-binding efficiency critically determines enzyme activity.Therefore, to enhance β-carotene hydroxylase's catalytic activity,reduce substrate accumulation, and promote astaxanthin biosynthesis from β-carotene,we performed an alanine scan on the pocket region, aiming to identity the critical amino acids affecting the enzyme activity. Several mutants with alanine replacement were generated.Sequentially,molecular docking was performed by CB-Dock2 to obtain the binding energy between substate and mutants.Based on the principle that higher binding ability between enzyme and substrate generally have lower binding energy,four mutants (ILE102ALA, SER96ALA, CYS191ALA, and THR213ALA) were subjected to astaxanthin productivity analysis (Figure 3a,b).We register them as new parts as HpCrtZILE102ALA(BBa_255CQOV8), HpCrtZSER96ALA(BBa_25YM5299), HpCrtZCYS191ALA(BBa_259A010E), and HpCrtZTHR213ALA(BBa_259A010E) .
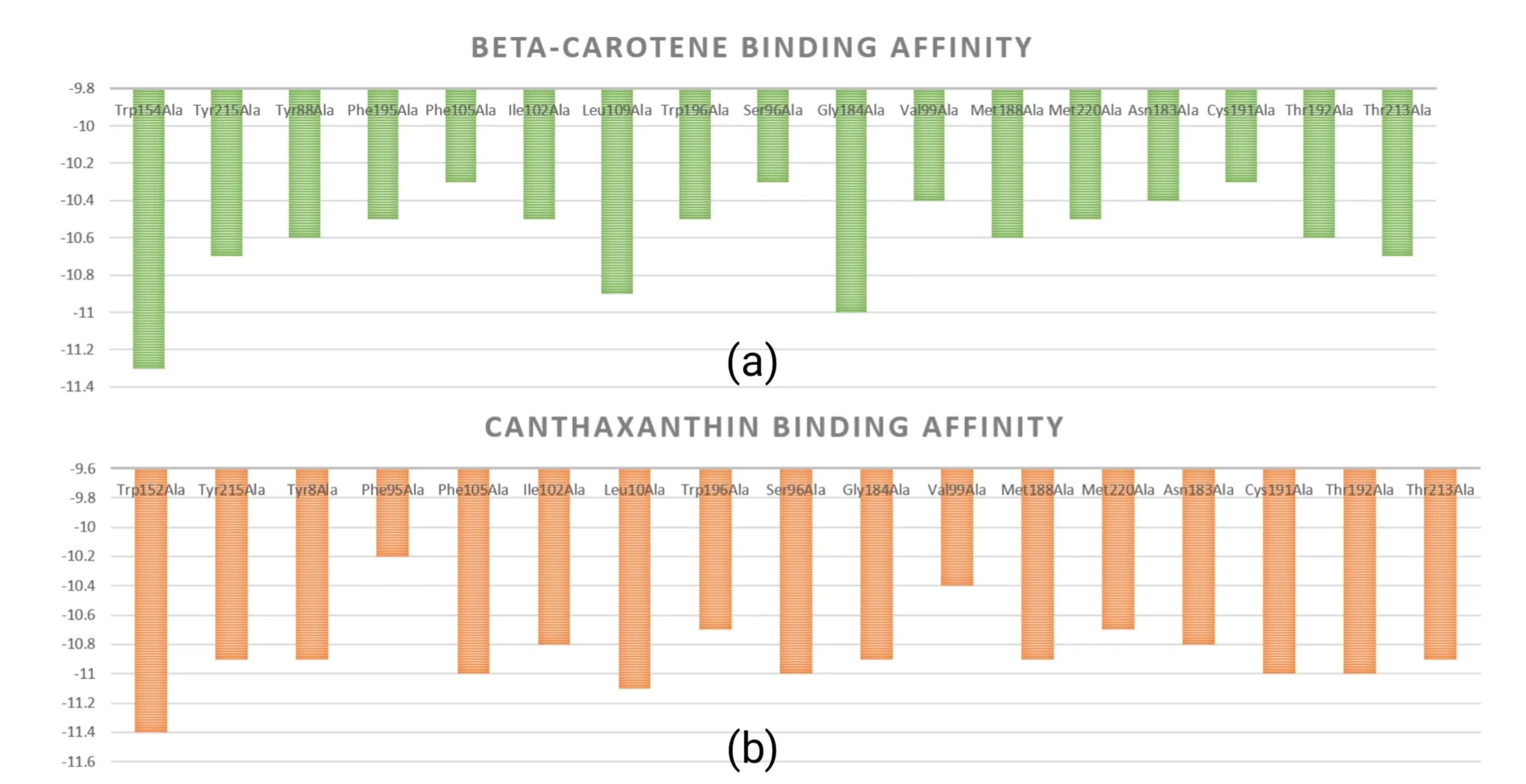 Figure 3. The binding energy between each mutant β-carotene hydroxylase with alanine replacement in the binding pocket and β-carotene (a)/canthaxanthin (b)
Figure 3. The binding energy between each mutant β-carotene hydroxylase with alanine replacement in the binding pocket and β-carotene (a)/canthaxanthin (b)
The amino acids of four mutants were subjected to AlphaFold2 to achieve their model files. Subsequently, their model files and substrate β-carotene were uploaded to the CB-Dock2 website where molecular docking simulations were performed. Results suggested that their Vina score were lower than -10, indicating their higher binding ability to β-carotene (Figure4-7).
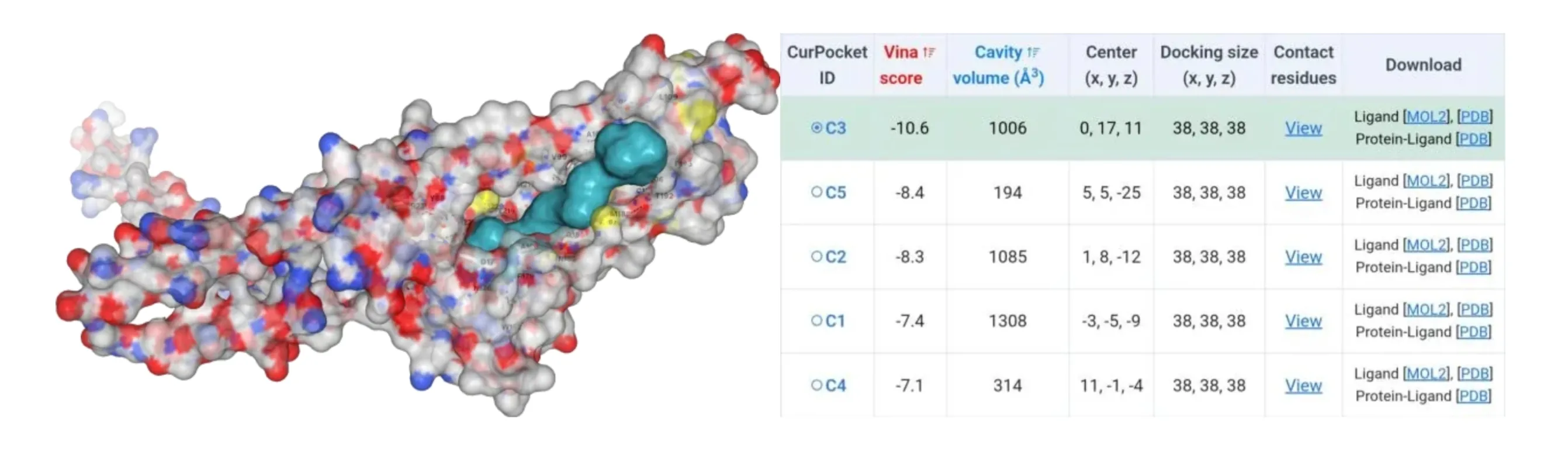 Figure 4.Molecular docking between HpCrtZILE102ALA mutant with β-carotene
Figure 4.Molecular docking between HpCrtZILE102ALA mutant with β-carotene
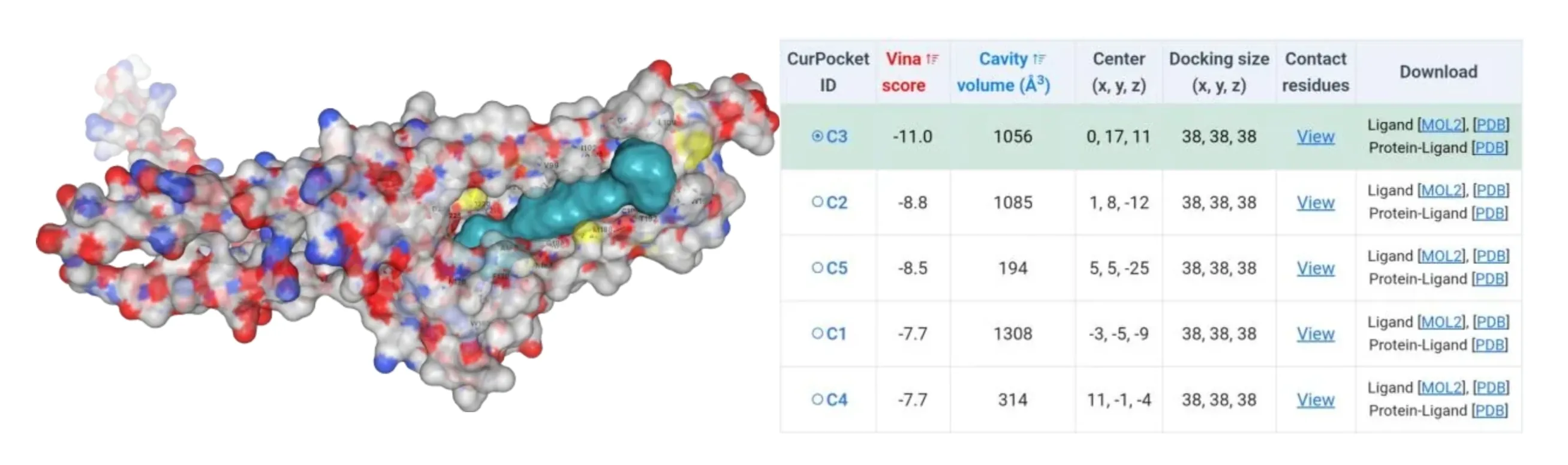 Figure 5.Molecular docking between HpCrtZSER102ALA mutant with β-carotene
Figure 5.Molecular docking between HpCrtZSER102ALA mutant with β-carotene
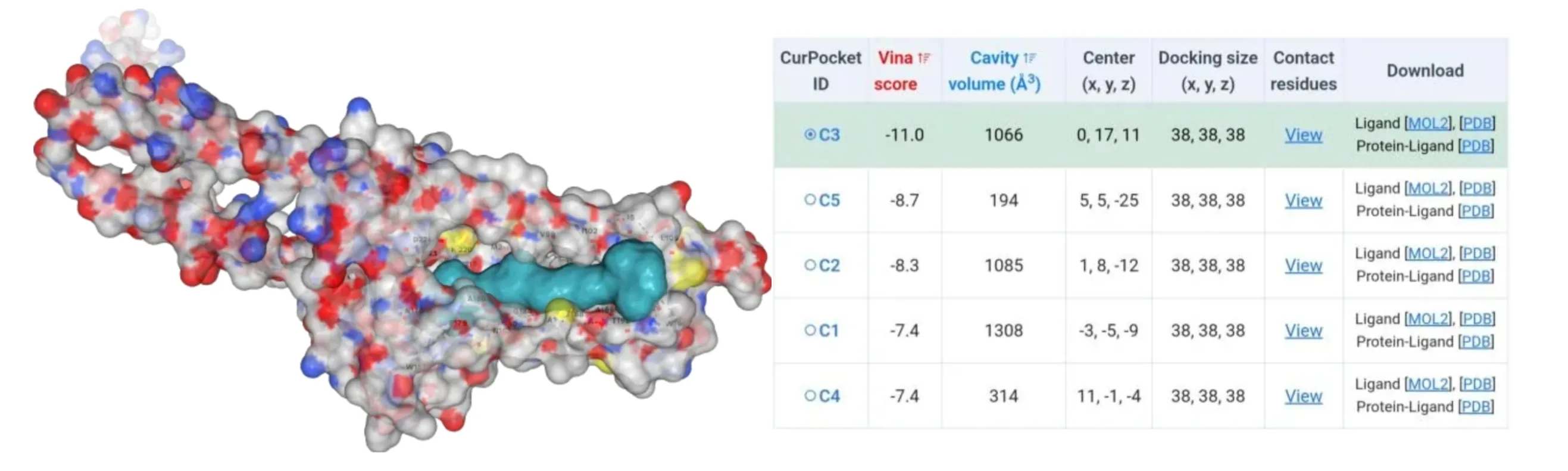 Figure 6.Molecular docking between HpCrtZCYS102ALA mutant with β-carotene
Figure 6.Molecular docking between HpCrtZCYS102ALA mutant with β-carotene
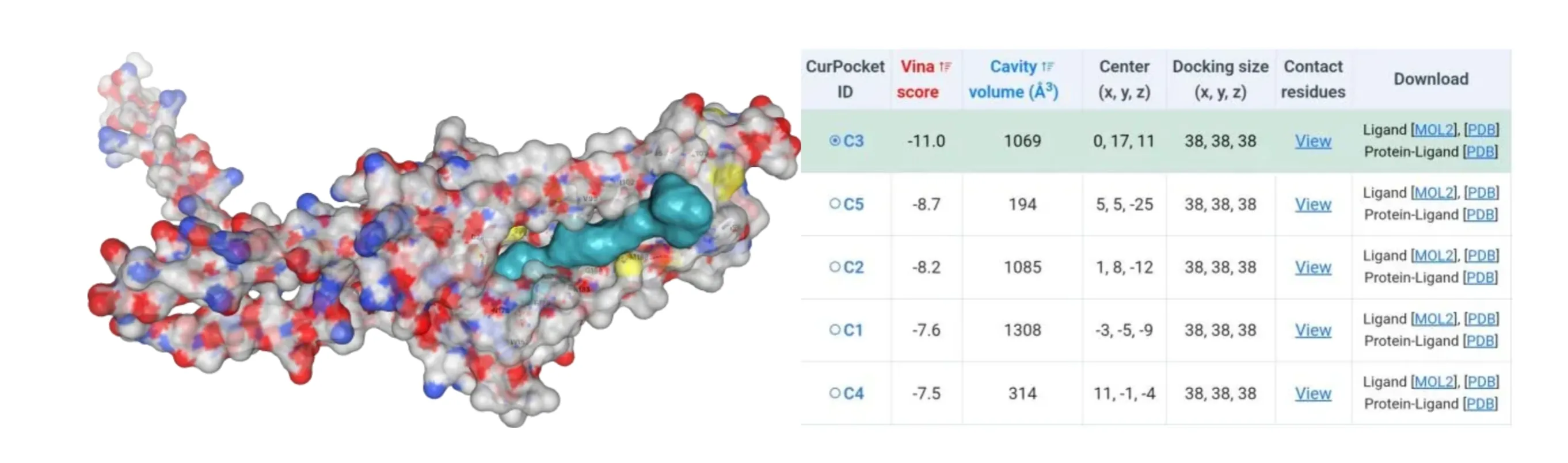 Figure 7.Molecular docking between HpCrtZTHR102ALA mutant with β-carotene
Figure 7.Molecular docking between HpCrtZTHR102ALA mutant with β-carotene
The amino acids of four mutants were subjected to AlphaFold2 to achieve their model files. Subsequently, their model files and substrate canthaxanthin were uploaded to the CB-Dock2 website where molecular docking simulations were performed. Except the Vina score for HpCrtZCYS102ALA has the Vina score of -9.4, the rest three have Vina score lower than -10 (Figure5-11).
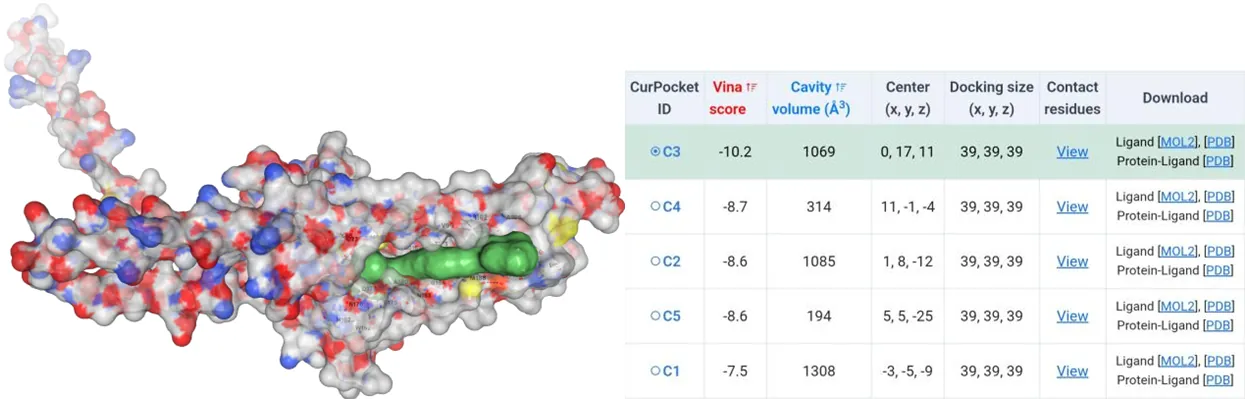 Figure 8.Molecular docking between HpCrtZILE102ALA mutant with canthaxanthin.
Figure 8.Molecular docking between HpCrtZILE102ALA mutant with canthaxanthin.
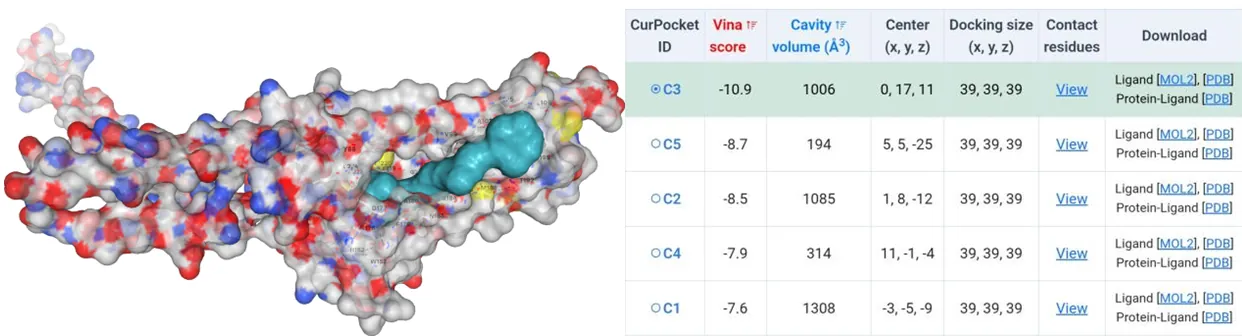 Figure 9.Molecular docking between HpCrtZSER102ALA mutant with canthaxanthin.
Figure 9.Molecular docking between HpCrtZSER102ALA mutant with canthaxanthin.
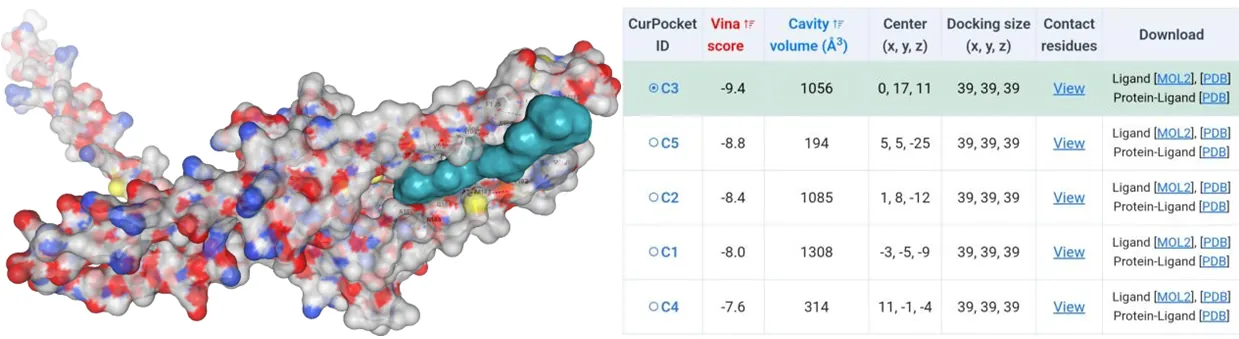 Figure 10.Molecular docking between HpCrtZCYS102ALA mutant with canthaxanthin.
Figure 10.Molecular docking between HpCrtZCYS102ALA mutant with canthaxanthin.
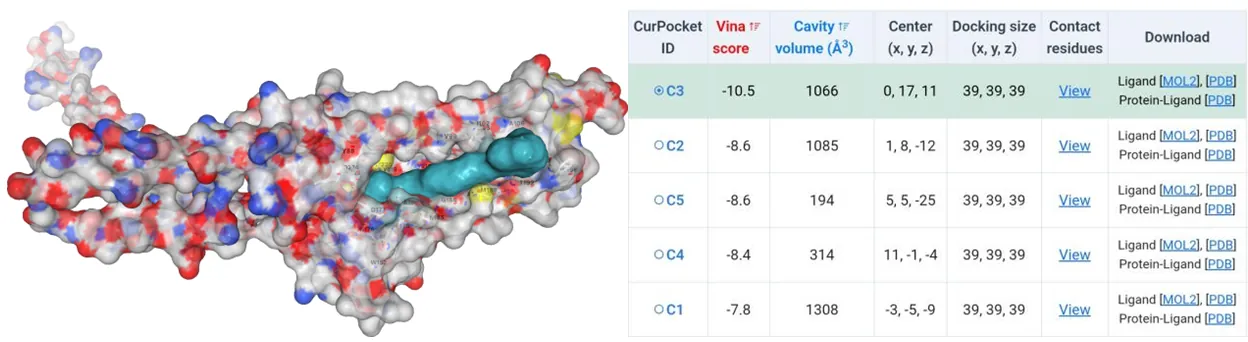 Figure 11.Molecular docking between HpCrtZTHR102ALA mutant with canthaxanthin.
Figure 11.Molecular docking between HpCrtZTHR102ALA mutant with canthaxanthin.
Based on the results obtained from molecular docking analysis, we selected four mutants including HpCrtZILE102ALA, HpCrtZSER96ALA, HpCrtZCYS191ALA, and HpCrtZTHR213ALA and tested their functions on the astaxanthin synthesis in E. coli. These mutants were obtained through chemical synthesis and then used to replace the HpCrtZ gene on the pET-ast plasmid, resulting in pET-astILE102ALA, pET-astSER96ALA, pET-astCYS191ALA, and pET-astTHR213ALA, respectively. The mutant plasmids were transformed into E. coli BL21(DE3) and the genes related with astaxanthin synthesis were expressed by IPTG induction. According to the HPLC results, all the mutant plasmids succeeding in producing astaxanthin, meaning that the methods we conducted site-directed mutagenesis in HpCrtZ based on the prediction of binding pocket were successful. Of these mutants, HpCrtZ (CYS191ALA, BBa_259A010E) showed the highest catalytic activity improving astaxanthin production by 1.28-fold (Figure 12). Our findings would help the people to increase astaxanthin production in the future.
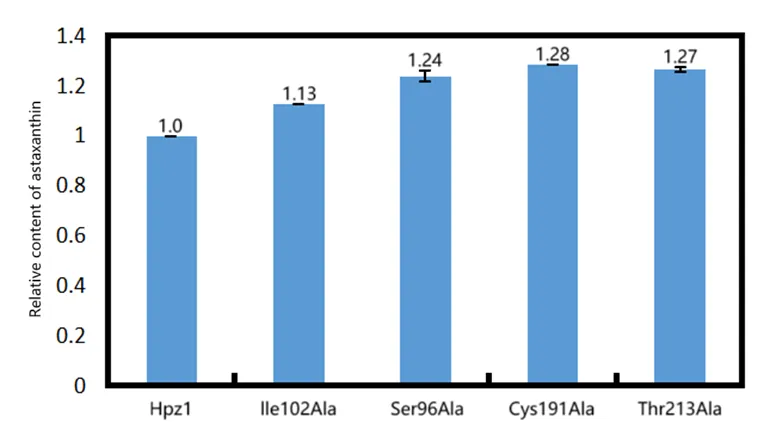 Figure 12. Effect of β-carotene hydroxylase mutants on astaxanthin production
Figure 12. Effect of β-carotene hydroxylase mutants on astaxanthin production
Learning more, please go to the Results page.
[1] Huang, D. Q., Liu, W. F., Li, A. G., Hu, Z. L., Wang, J. X. & Wang, C. G. Cloning and identification of a novel β-carotene hydroxylase gene from Haematococcus pluvialis and its function in Escherichia coli. Algal Res. 55, 102245 (2021).
[2] Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
[3] Liu, Y., Yang, X., Gan, J., Cao, Y. et al. CB-Dock2: improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 50, W159–W167 (2022).